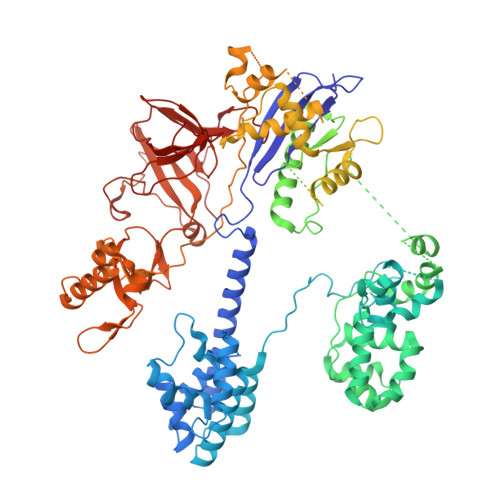
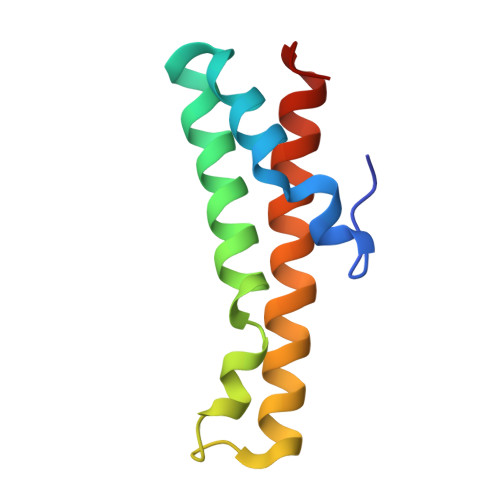
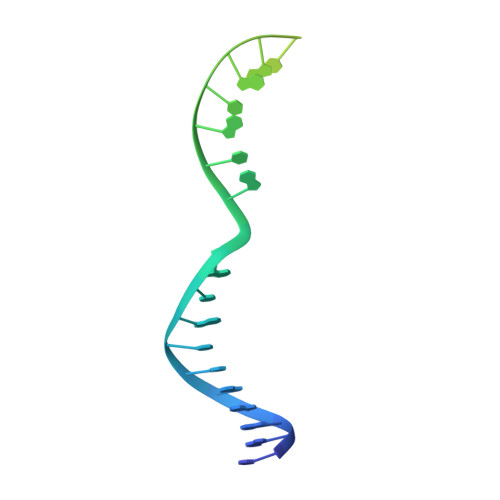
AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Sun, W., Cheng, Z., Wang, J., Yang, J., Li, X., Wang, J., Chen, M., Yang, X., Sheng, G., Lou, J., Wang, Y.(2023) Proc Natl Acad Sci U S A 120: e2303675120-e2303675120
- PubMed: 37494395
- DOI: https://doi.org/10.1073/pnas.2303675120
- Primary Citation of Related Structures:
8HNS, 8HNT, 8HNV, 8HNW - PubMed Abstract:
Anti-CRISPR (Acr) proteins are encoded by phages and other mobile genetic elements and inhibit host CRISPR-Cas immunity using versatile strategies. AcrIIC4 is a broad-spectrum Acr that inhibits the type II-C CRISPR-Cas9 system in several species by an unknown mechanism. Here, we determined a series of structures of Haemophilus parainfluenzae Cas9 (HpaCas9)-sgRNA in complex with AcrIIC4 and/or target DNA, as well as the crystal structure of AcrIIC4 alone. We found that AcrIIC4 resides in the crevice between the REC1 and REC2 domains of HpaCas9, where its extensive interactions restrict the mobility of the REC2 domain and prevent the unwinding of target double-stranded (ds) DNA at the PAM-distal end. Therefore, the full-length guide RNA:target DNA heteroduplex fails to form in the presence of AcrIIC4, preventing Cas9 nuclease activation. Altogether, our structural and biochemical studies illuminate a unique Acr mechanism that allows DNA binding to the Cas9 effector complex but blocks its cleavage by preventing R-loop formation, a key step supporting DNA cleavage by Cas9.
- Key Laboratory of RNA Biology, Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: