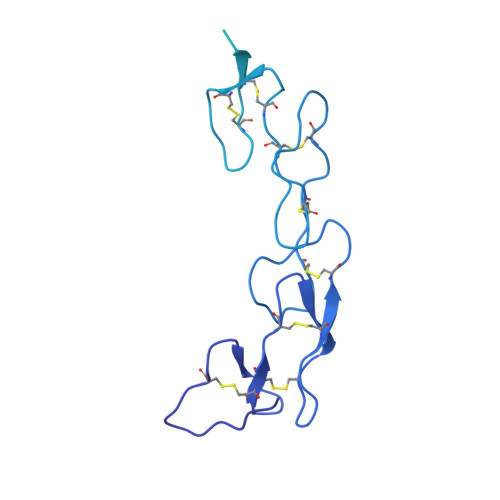
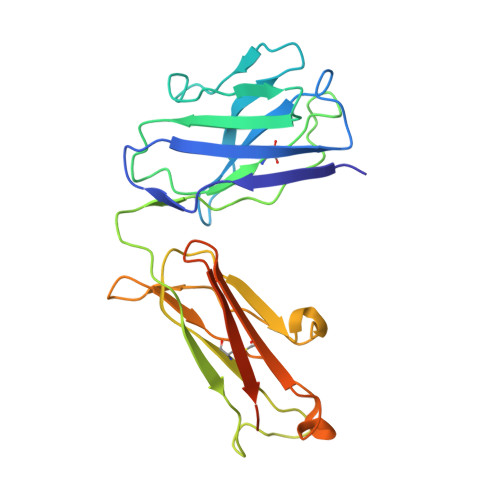
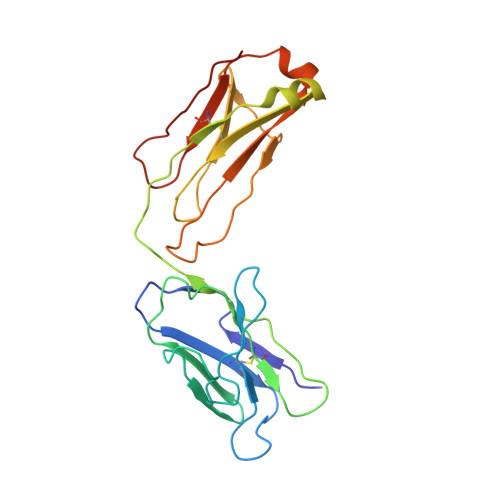
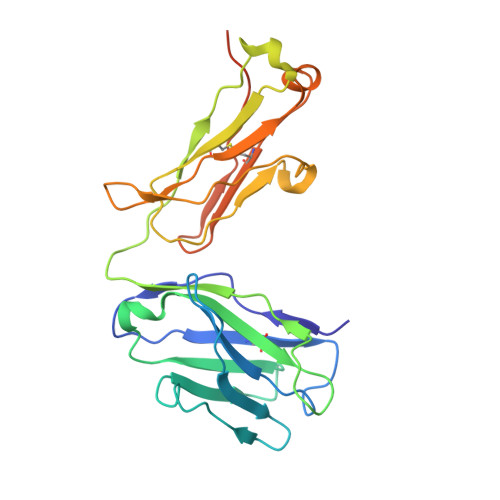
Development of a 1:1-binding biparatopic anti-TNFR2 antagonist by reducing signaling activity through epitope selection.
Akiba, H., Fujita, J., Ise, T., Nishiyama, K., Miyata, T., Kato, T., Namba, K., Ohno, H., Kamada, H., Nagata, S., Tsumoto, K.(2023) Commun Biol 6: 987-987
- PubMed: 37758868
- DOI: https://doi.org/10.1038/s42003-023-05326-8
- Primary Citation of Related Structures:
8HLB - PubMed Abstract:
Conventional bivalent antibodies against cell surface receptors often initiate unwanted signal transduction by crosslinking two antigen molecules. Biparatopic antibodies (BpAbs) bind to two different epitopes on the same antigen, thus altering crosslinking ability. In this study, we develop BpAbs against tumor necrosis factor receptor 2 (TNFR2), which is an attractive immune checkpoint target. Using different pairs of antibody variable regions specific to topographically distinct TNFR2 epitopes, we successfully regulate the size of BpAb-TNFR2 immunocomplexes to result in controlled agonistic activities. Our series of results indicate that the relative positions of the two epitopes recognized by the BpAb are critical for controlling its signaling activity. One particular antagonist, Bp109-92, binds TNFR2 in a 1:1 manner without unwanted signal transduction, and its structural basis is determined using cryo-electron microscopy. This antagonist suppresses the proliferation of regulatory T cells expressing TNFR2. Therefore, the BpAb format would be useful in designing specific and distinct antibody functions.
- Graduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto, 606-8501, Japan. hakiba@pharm.kyoto-u.ac.jp.
Organizational Affiliation: