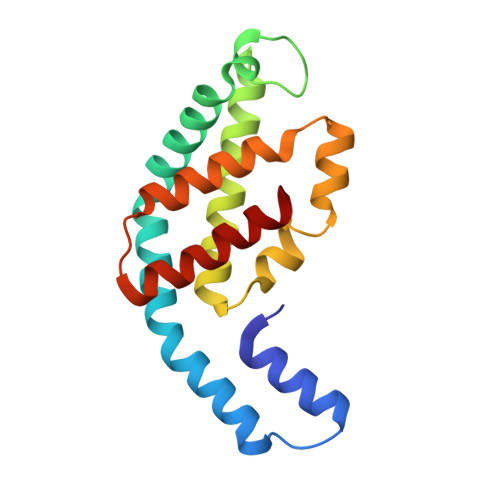
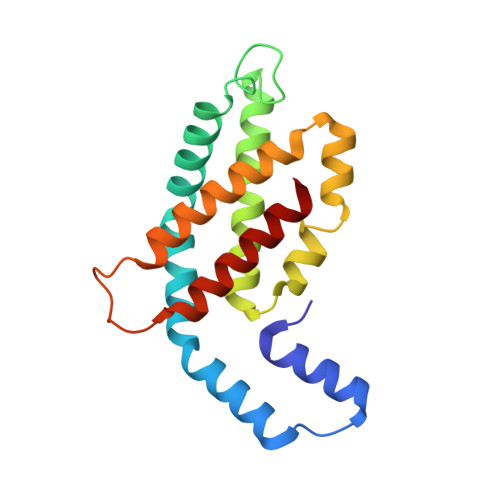
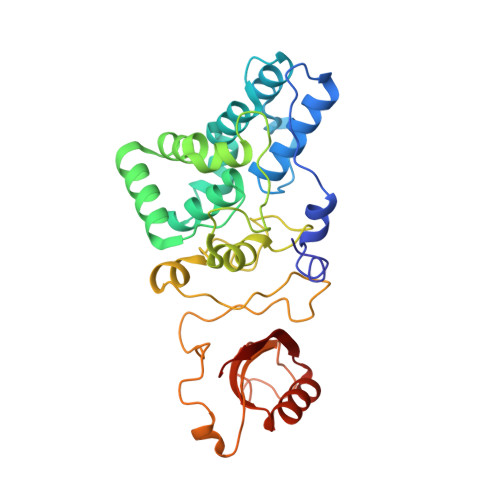
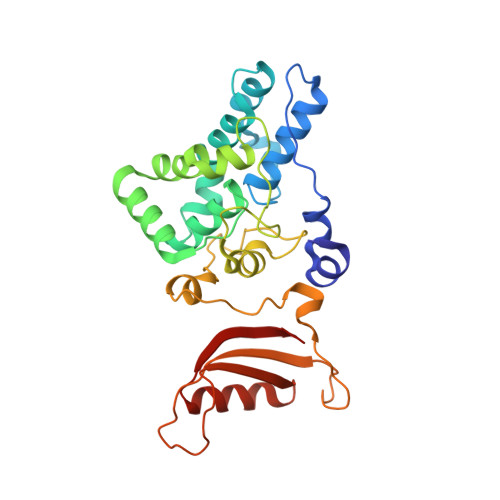
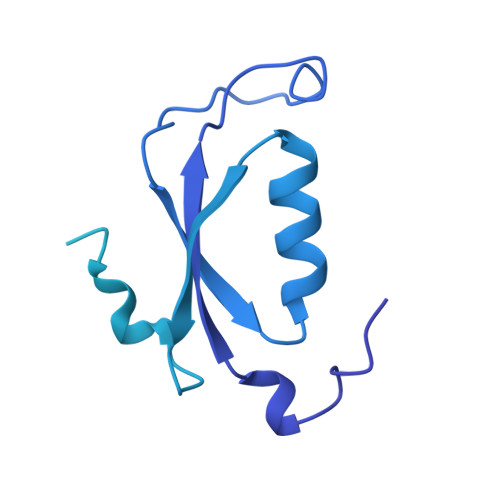
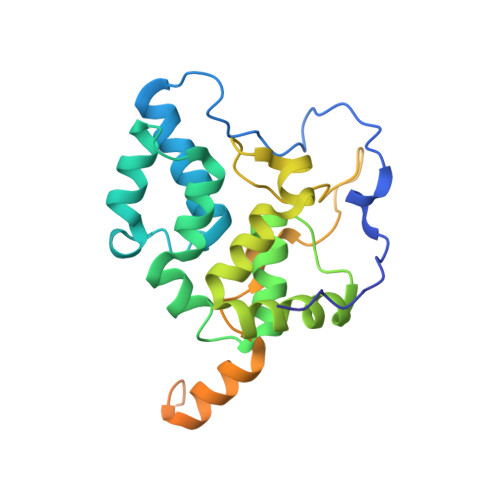
Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Zheng, L., Zhang, Z., Wang, H., Zheng, Z., Wang, J., Liu, H., Chen, H., Dong, C., Wang, G., Weng, Y., Gao, N., Zhao, J.(2023) Nat Commun 14: 3961-3961
- PubMed: 37407580
- DOI: https://doi.org/10.1038/s41467-023-39689-7
- Primary Citation of Related Structures:
8HFQ - PubMed Abstract:
Phycobilisomes (PBS) are the major light harvesting complexes of photosynthesis in the cyanobacteria and red algae. CpcL-PBS is a type of small PBS in cyanobacteria that transfers energy directly to photosystem I without the core structure. Here we report the cryo-EM structure of the CpcL-PBS from the cyanobacterium Synechocystis sp. PCC 6803 at 2.6-Å resolution. The structure shows the CpcD domain of ferredoxin: NADP + oxidoreductase is located at the distal end of CpcL-PBS, responsible for its attachment to PBS. With the evidence of ultrafast transient absorption and fluorescence spectroscopy, the roles of individual bilins in energy transfer are revealed. The bilin 1I β 82 2 located near photosystem I has an enhanced planarity and is the red-bilin responsible for the direct energy transfer to photosystem I.
- School of Life Sciences, Peking University, Beijing, 100871, China.
Organizational Affiliation: