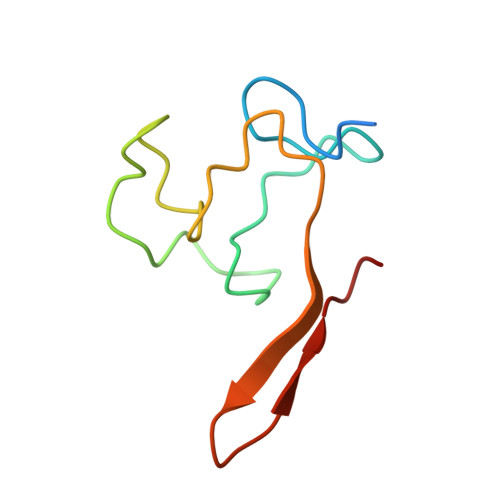
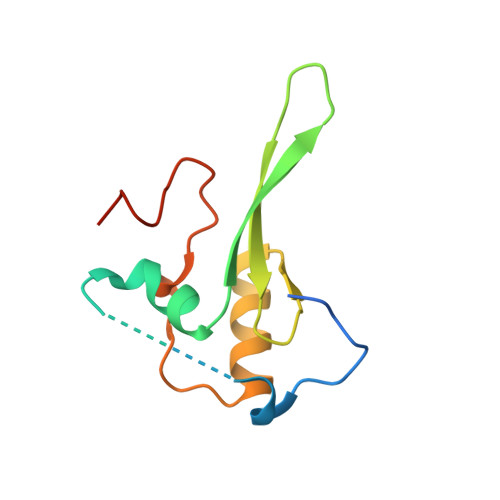
Structural evidence for protein-protein interaction between the non-canonical methyl-CpG-binding domain of SETDB proteins and C11orf46.
Mahana, Y., Ariyoshi, M., Nozawa, R.S., Shibata, S., Nagao, K., Obuse, C., Shirakawa, M.(2024) Structure 32: 304-315.e5
- PubMed: 38159574
- DOI: https://doi.org/10.1016/j.str.2023.12.001
- Primary Citation of Related Structures:
8HFP - PubMed Abstract:
SETDB1 and SETDB2 mediate trimethylation of histone H3 lysine 9 (H3K9), an epigenetic hallmark of repressive chromatin. They contain a non-canonical methyl-CpG-binding domain (MBD) and bifurcated SET domain, implying interplay between H3K9 trimethylation and DNA methylation in SETDB functions. Here, we report the crystal structure of human SETDB2 MBD bound to the cysteine-rich domain of a zinc-binding protein, C11orf46. SETDB2 MBD comprises the conserved MBD core and a unique N-terminal extension. Although the MBD core has the conserved basic concave surface for DNA binding, it utilizes it for recognition of the cysteine-rich domain of C11orf46. This interaction involves the conserved arginine finger motif and the unique N-terminal extension of SETDB2 MBD, with a contribution from intermolecular β-sheet formation. Thus, the non-canonical MBD of SETDB1/2 seems to have lost methylated DNA-binding ability but gained a protein-protein interaction surface. Our findings provide insight into the molecular assembly of SETDB-associated repression complexes.
- Department of Molecular Engineering, Graduate School of Engineering, Kyoto University, Nishikyo-Ku, Kyoto 615-8510, Japan.
Organizational Affiliation: