Cryo-Electron Microscopy Structure of the Mycobacterium tuberculosi s Cytochrome bcc : aa 3 Supercomplex and a Novel Inhibitor Targeting Subunit Cytochrome c I.
Mathiyazakan, V., Wong, C.F., Harikishore, A., Pethe, K., Gruber, G.(2023) Antimicrob Agents Chemother 67: e0153122-e0153122
- PubMed: 37158740
- DOI: https://doi.org/10.1128/aac.01531-22
- Primary Citation of Related Structures:
8HCR - PubMed Abstract:
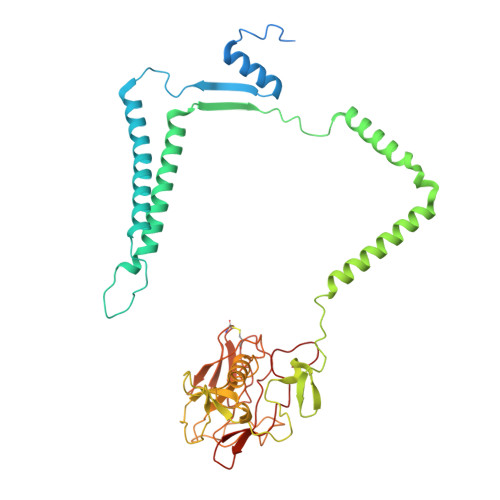
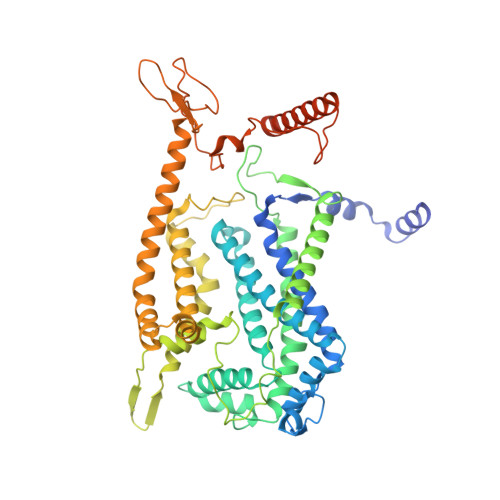
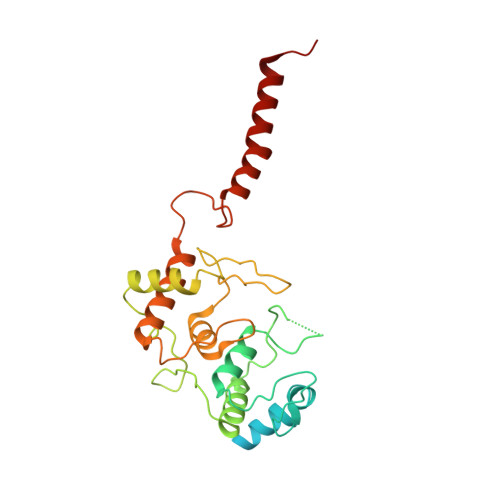
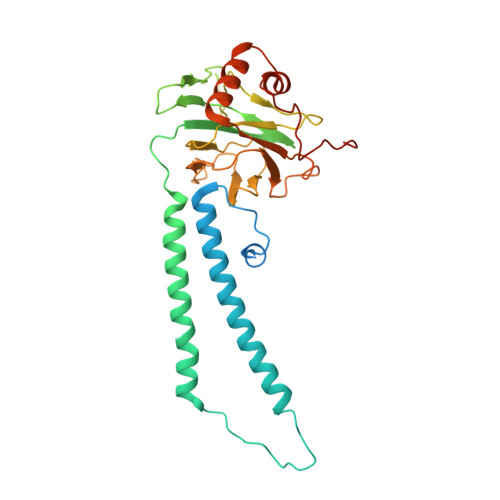
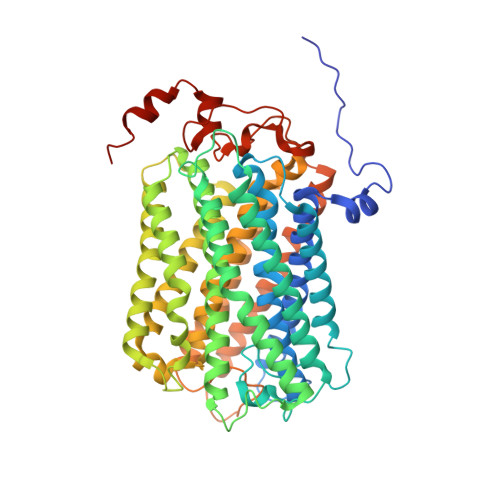
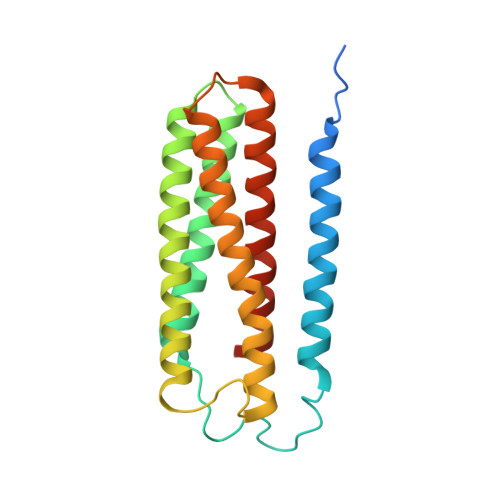
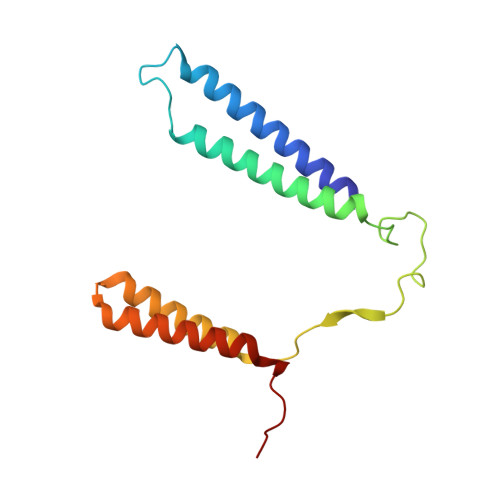
The mycobacterial cytochrome bcc:aa 3 complex deserves the name "supercomplex" since it combines three cytochrome oxidases-cytochrome bc , cytochrome c , and cytochrome aa 3 -into one supramolecular machine and performs electron transfer for the reduction of oxygen to water and proton transport to generate the proton motive force for ATP synthesis. Thus, the bcc:aa 3 complex represents a valid drug target for Mycobacterium tuberculosis infections. The production and purification of an entire M. tuberculosis cytochrome bcc:aa 3 are fundamental for biochemical and structural characterization of this supercomplex, paving the way for new inhibitor targets and molecules. Here, we produced and purified the entire and active M. tuberculosis cyt- bcc:aa 3 oxidase, as demonstrated by the different heme spectra and an oxygen consumption assay. The resolved M. tuberculosis cyt- bcc:aa 3 cryo-electron microscopy structure reveals a dimer with its functional domains involved in electron, proton, oxygen transfer, and oxygen reduction. The structure shows the two cytochrome c I c II head domains of the dimer, the counterpart of the soluble mitochondrial cytochrome c , in a so-called "closed state," in which electrons are translocated from the bcc to the aa 3 domain. The structural and mechanistic insights provided the basis for a virtual screening campaign that identified a potent M. tuberculosis cyt- bcc:aa 3 inhibitor, cyt Mycc 1. cyt Mycc 1 targets the mycobacterium-specific α3-helix of cytochrome c I and interferes with oxygen consumption by interrupting electron translocation via the c I c II head. The successful identification of a new cyt- bcc:aa 3 inhibitor demonstrates the potential of a structure-mechanism-based approach for novel compound development.
- Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Republic of Singapore.
Organizational Affiliation: