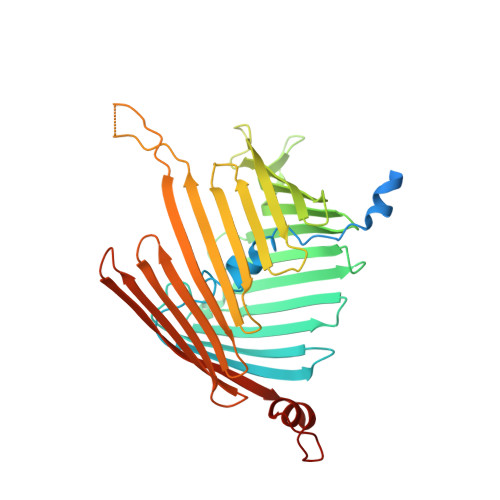
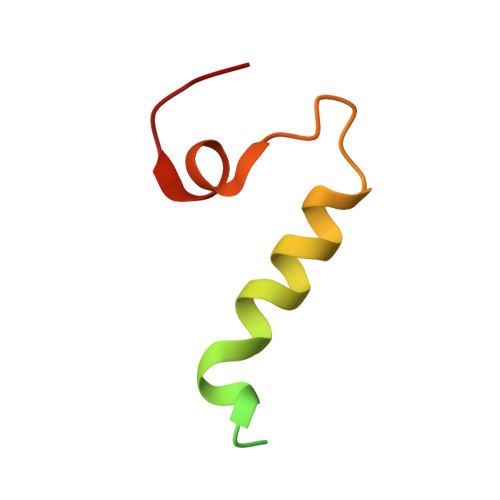
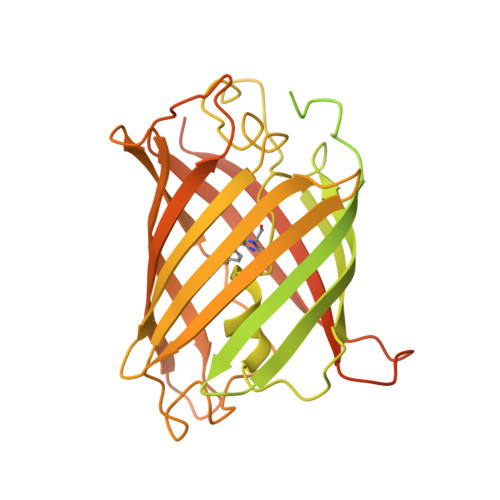
Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.
Zhou, X., Yang, Y., Wang, G., Wang, S., Sun, D., Ou, X., Lian, Y., Li, L.(2023) Nat Struct Mol Biol 30: 1996-2008
- PubMed: 37696957
- DOI: https://doi.org/10.1038/s41594-023-01103-7
- Primary Citation of Related Structures:
8HCO - PubMed Abstract:
Over half of mitochondrial proteins are imported from the cytosol via the pre-sequence pathway, controlled by the TOM complex in the outer membrane and the TIM23 complex in the inner membrane. The mechanisms through which proteins are translocated via the TOM and TIM23 complexes remain unclear. Here we report the assembly of the active TOM-TIM23 supercomplex of Saccharomyces cerevisiae with translocating polypeptide substrates. Electron cryo-microscopy analyses reveal that the polypeptide substrates pass the TOM complex through the center of a Tom40 subunit, interacting with a glutamine-rich region. Structural and biochemical analyses show that the TIM23 complex contains a heterotrimer of the subunits Tim23, Tim17 and Mgr2. The polypeptide substrates are shielded from lipids by Mgr2 and Tim17, which creates a translocation pathway characterized by a negatively charged entrance and a central hydrophobic region. These findings reveal an unexpected pre-sequence pathway through the TOM-TIM23 supercomplex spanning the double membranes of mitochondria.
- State Key Laboratory of Membrane Biology, Peking-Tsinghua Center for Life Sciences, School of Life Sciences, Peking University, Beijing, China.
Organizational Affiliation: