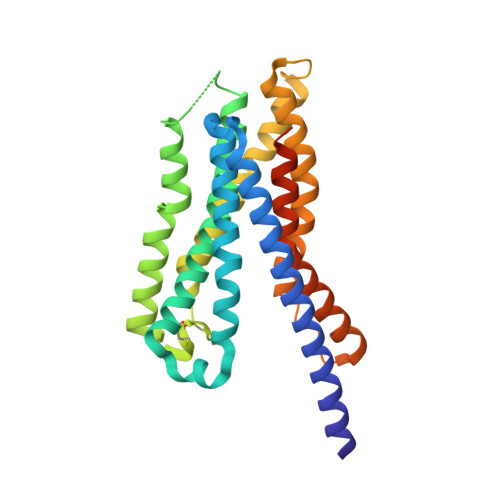
Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Kim, H., Lim, T., Ha, G.E., Lee, J.Y., Kim, J.W., Chang, N., Kim, S.H., Kim, K.H., Lee, J., Cho, Y., Kim, B.W., Abrahamsson, A., Kim, S.H., Kim, H.J., Park, S., Lee, S.J., Park, J., Cheong, E., Kim, B.M., Cho, H.S.(2023) Exp Mol Med 55: 2039-2050
- PubMed: 37653040
- DOI: https://doi.org/10.1038/s12276-023-01082-1
- Primary Citation of Related Structures:
8GTG, 8GTI, 8GTM - PubMed Abstract:
Thus far, attempts to develop drugs that target corticotropin-releasing hormone receptor 1 (CRF 1 R), a drug target in stress-related therapy, have been unsuccessful. Studies have focused on using high-resolution G protein-coupled receptor (GPCR) structures to develop drugs. X-ray free-electron lasers (XFELs), which prevent radiation damage and provide access to high-resolution compositions, have helped accelerate GPCR structural studies. We elucidated the crystal structure of CRF 1 R complexed with a BMK-I-152 antagonist at 2.75 Å using fixed-target serial femtosecond crystallography. The results revealed that two unique hydrogen bonds are present in the hydrogen bond network, the stalk region forms an alpha helix and the hydrophobic network contains an antagonist binding site. We then developed two antagonists-BMK-C203 and BMK-C205-and determined the CRF 1 R/BMK-C203 and CRF 1 R/BMK-C205 complex structures at 2.6 and 2.2 Å, respectively. BMK-C205 exerted significant antidepressant effects in mice and, thus, may be utilized to effectively identify structure-based drugs against CRF 1 R.
- Department of Systems Biology, College of Life Science and Biotechnology, Yonsei University, 50 Yonsei-ro, Seoul, 03722, Republic of Korea.
Organizational Affiliation: