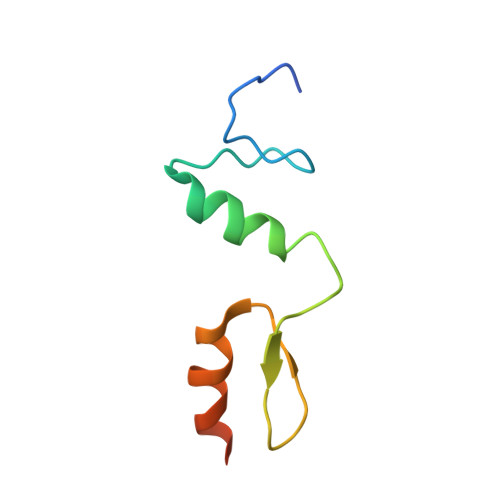
Structural insights into the recognition of telomeric variant repeat TTGGGG by broad-complex, tramtrack and bric-a-brac - zinc finger protein ZBTB10.
Wang, S., Xu, Z., Li, M., Lv, M., Shen, S., Shi, Y., Li, F.(2023) J Biological Chem 299: 102918-102918
- PubMed: 36657642
- DOI: https://doi.org/10.1016/j.jbc.2023.102918
- Primary Citation of Related Structures:
8GN3, 8GN4 - PubMed Abstract:
Multiple proteins bind to telomeric DNA and are important for the role of telomeres in genome stability. A recent study established a broad-complex, tramtrack and bric-à-brac - zinc finger (BTB-ZF) protein, ZBTB10 (zinc finger and BTB domain-containing protein 10), as a telomeric variant repeat-binding protein at telomeres that use an alternative method for lengthening telomeres). ZBTB10 specifically interacts with the double-stranded telomeric variant repeat sequence TTGGGG by employing its tandem C2H2 zinc fingers (ZF1-2). Here, we solved the crystal structure of human ZBTB10 ZF1-2 in complex with a double-stranded DNA duplex containing the sequence TTGGGG to assess the molecular details of this interaction. Combined with calorimetric analysis, we identified the vital residues in TTGGGG recognition and determined the specific recognition mechanisms that are different from those of TZAP (telomere zinc finger-associated protein), a recently defined telomeric DNA-binding protein. Following these studies, we further identified a single amino-acid mutant (Arg767Gln) of ZBTB10 ZF1-2 that shows a preference for the telomeric DNA repeat TTAGGG sequence. We solved the cocrystal structure, providing a structural basis for telomeric DNA recognition by C2H2 ZF proteins.
- MOE Key Laboratory for Cellular Dynamics, The School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, China.
Organizational Affiliation: