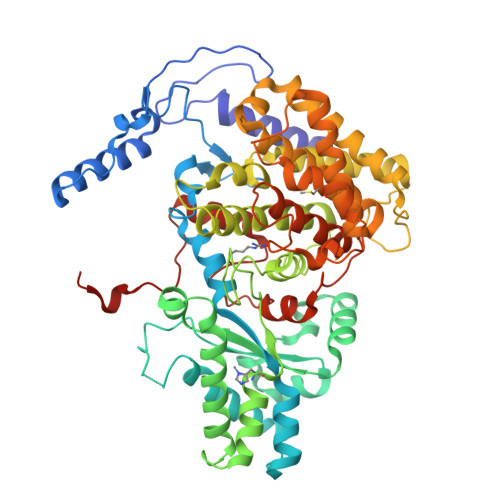
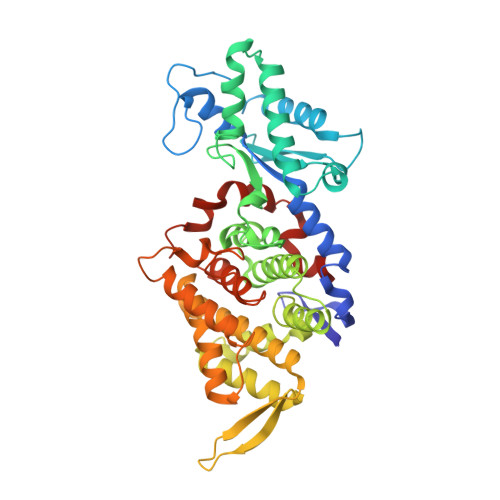
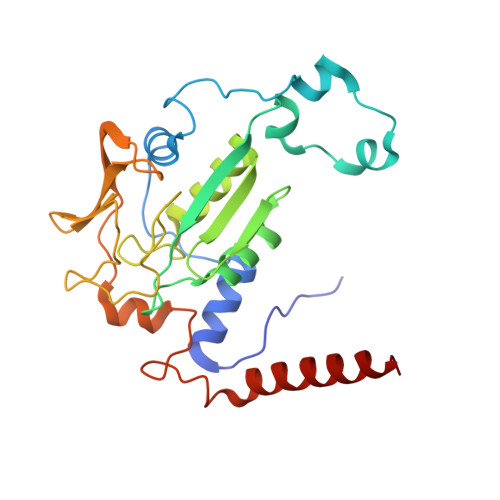
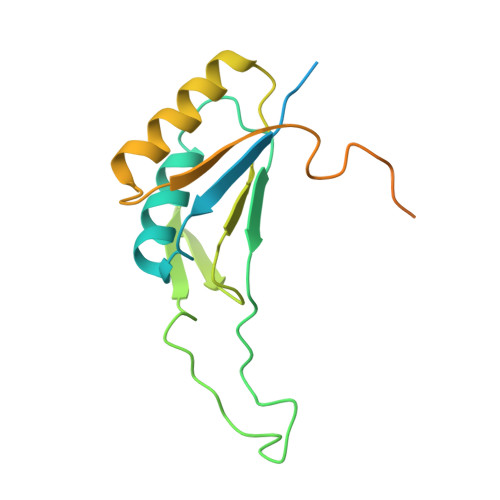
McrD binds asymmetrically to methyl-coenzyme M reductase improving active-site accessibility during assembly.
Chadwick, G.L., Joiner, A.M.N., Ramesh, S., Mitchell, D.A., Nayak, D.D.(2023) Proc Natl Acad Sci U S A 120: e2302815120-e2302815120
- PubMed: 37307484
- DOI: https://doi.org/10.1073/pnas.2302815120
- Primary Citation of Related Structures:
8GF5, 8GF6 - PubMed Abstract:
Methyl-coenzyme M reductase (MCR) catalyzes the formation of methane, and its activity accounts for nearly all biologically produced methane released into the atmosphere. The assembly of MCR is an intricate process involving the installation of a complex set of posttranslational modifications and the unique Ni-containing tetrapyrrole called coenzyme F 430 . Despite decades of research, details of MCR assembly remain largely unresolved. Here, we report the structural characterization of MCR in two intermediate states of assembly. These intermediate states lack one or both F 430 cofactors and form complexes with the previously uncharacterized McrD protein. McrD is found to bind asymmetrically to MCR, displacing large regions of the alpha subunit and increasing active-site accessibility for the installation of F 430 -shedding light on the assembly of MCR and the role of McrD therein. This work offers crucial information for the expression of MCR in a heterologous host and provides targets for the design of MCR inhibitors.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720.
Organizational Affiliation: