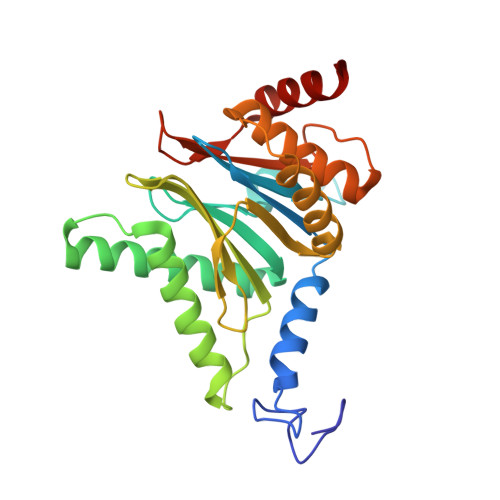
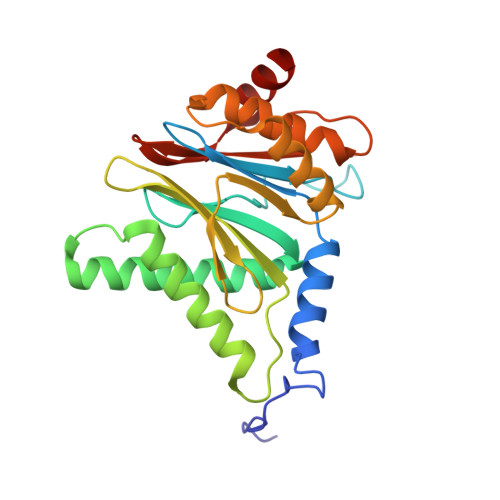
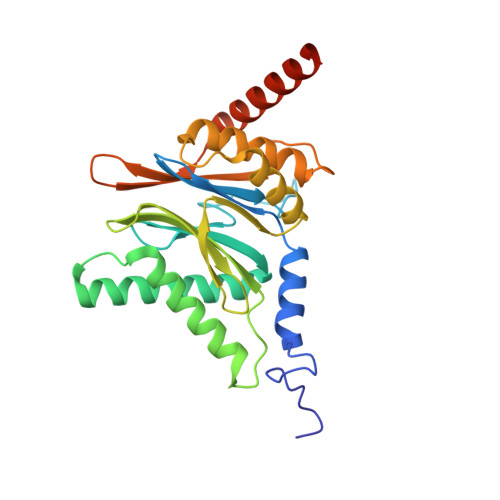
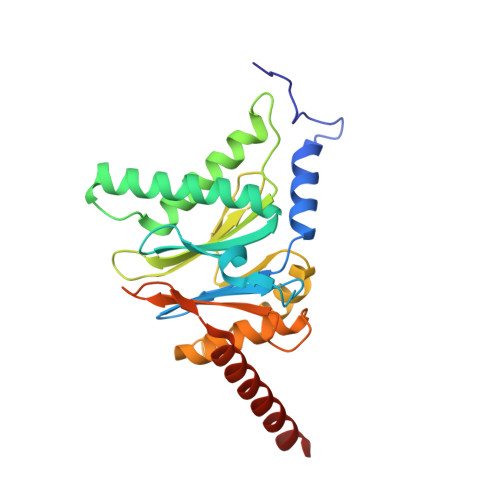
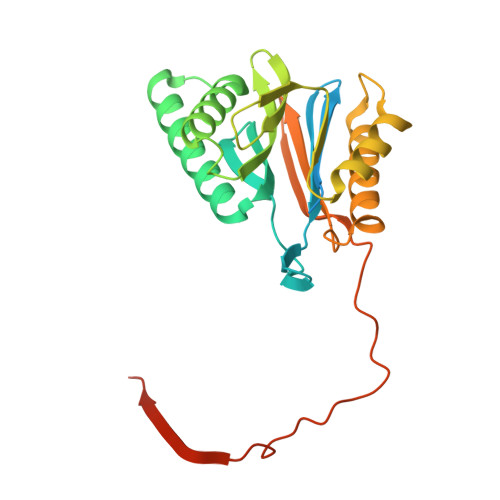
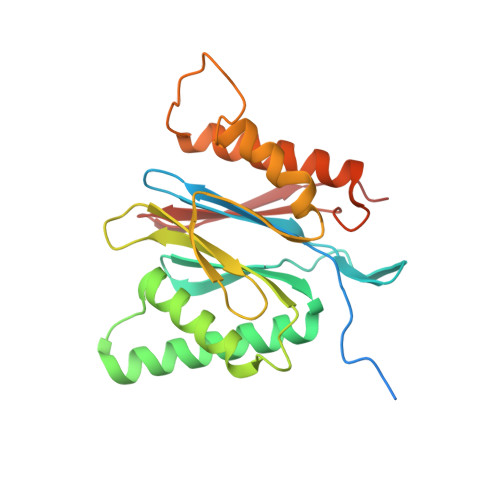
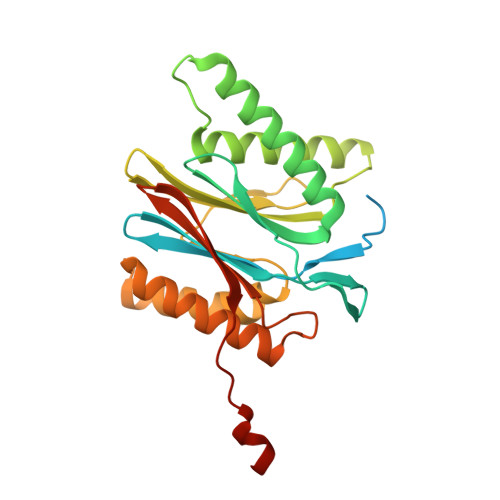
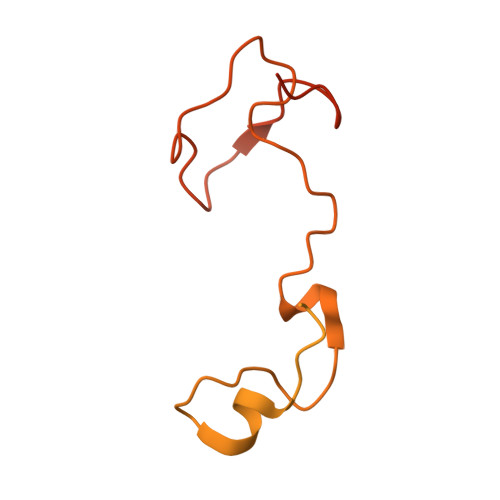
Eta igh-resolution structure of mammalian PI31-20S proteasome complex reveals mechanism of proteasome inhibition.
Hsu, H.C., Wang, J., Kjellgren, A., Li, H., DeMartino, G.N.(2023) J Biological Chem 299: 104862-104862
- PubMed: 37236357
- DOI: https://doi.org/10.1016/j.jbc.2023.104862
- Primary Citation of Related Structures:
8FZ5, 8FZ6 - PubMed Abstract:
Proteasome-catalyzed protein degradation mediates and regulates critical aspects of many cellular functions and is an important element of proteostasis in health and disease. Proteasome function is determined in part by the types of proteasome holoenzymes formed between the 20S core particle that catalyzes peptide bond hydrolysis and any of multiple regulatory proteins to which it binds. One of these regulators, PI31, was previously identified as an in vitro 20S proteasome inhibitor, but neither the molecular mechanism nor the possible physiologic significance of PI31-mediated proteasome inhibition has been clear. Here we report a high-resolution cryo-EM structure of the mammalian 20S proteasome in complex with PI31. The structure shows that two copies of the intrinsically disordered carboxyl terminus of PI31 are present in the central cavity of the closed-gate conformation of the proteasome and interact with proteasome catalytic sites in a manner that blocks proteolysis of substrates but resists their own degradation. The two inhibitory polypeptide chains appear to originate from PI31 monomers that enter the catalytic chamber from opposite ends of the 20S cylinder. We present evidence that PI31 can inhibit proteasome activity in mammalian cells and may serve regulatory functions for the control of cellular proteostasis.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, Michigan, USA.
Organizational Affiliation: