Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Holt, G.T., Gorman, J., Wang, S., Lowegard, A.U., Zhang, B., Liu, T., Lin, B.C., Louder, M.K., Frenkel, M.S., McKee, K., O'Dell, S., Rawi, R., Shen, C.H., Doria-Rose, N.A., Kwong, P.D., Donald, B.R.(2023) Cell Rep 42: 112711-112711
- PubMed: 37436900
- DOI: https://doi.org/10.1016/j.celrep.2023.112711
- Primary Citation of Related Structures:
8FK5, 8FL1, 8FLW - PubMed Abstract:
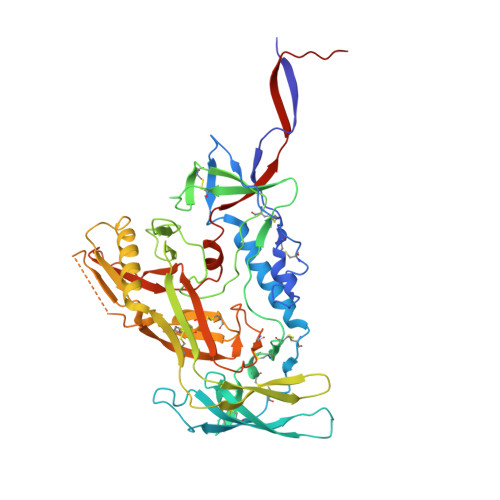
Broadly neutralizing antibodies (bNAbs) against HIV can reduce viral transmission in humans, but an effective therapeutic will require unusually high breadth and potency of neutralization. We employ the OSPREY computational protein design software to engineer variants of two apex-directed bNAbs, PGT145 and PG9RSH, resulting in increases in potency of over 100-fold against some viruses. The top designed variants improve neutralization breadth from 39% to 54% at clinically relevant concentrations (IC 80 < 1 μg/mL) and improve median potency (IC 80 ) by up to 4-fold over a cross-clade panel of 208 strains. To investigate the mechanisms of improvement, we determine cryoelectron microscopy structures of each variant in complex with the HIV envelope trimer. Surprisingly, we find the largest increases in breadth to be a result of optimizing side-chain interactions with highly variable epitope residues. These results provide insight into mechanisms of neutralization breadth and inform strategies for antibody design and improvement.
- Department of Computer Science, Duke University, Durham, NC, USA; Program in Computational Biology & Bioinformatics, Duke University, Durham, NC, USA.
Organizational Affiliation: