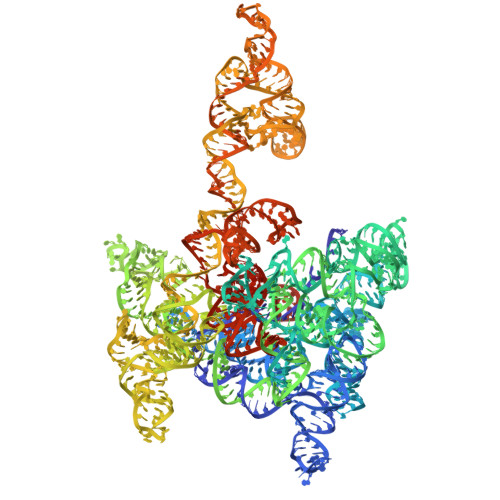
Structural basis of branching during RNA splicing.
Haack, D.B., Rudolfs, B., Zhang, C., Lyumkis, D., Toor, N.(2024) Nat Struct Mol Biol 31: 179-189
- PubMed: 38057551
- DOI: https://doi.org/10.1038/s41594-023-01150-0
- Primary Citation of Related Structures:
8FLI - PubMed Abstract:
Branching is a critical step in RNA splicing that is essential for 5' splice site selection. Recent spliceosome structures have led to competing models for the recognition of the invariant adenosine at the branch point. However, there are no structures of any splicing complex with the adenosine nucleophile docked in the active site and positioned to attack the 5' splice site. Thus we lack a mechanistic understanding of adenosine selection and splice site recognition during RNA splicing. Here we present a cryo-electron microscopy structure of a group II intron that reveals that active site dynamics are coupled to the formation of a base triple within the branch-site helix that positions the 2'-OH of the adenosine for nucleophilic attack on the 5' scissile phosphate. This structure, complemented with biochemistry and comparative analyses to splicing complexes, supports a base triple model of adenosine recognition for branching within group II introns and the evolutionarily related spliceosome.
- Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA, USA. dhaack@ucsd.edu.
Organizational Affiliation: