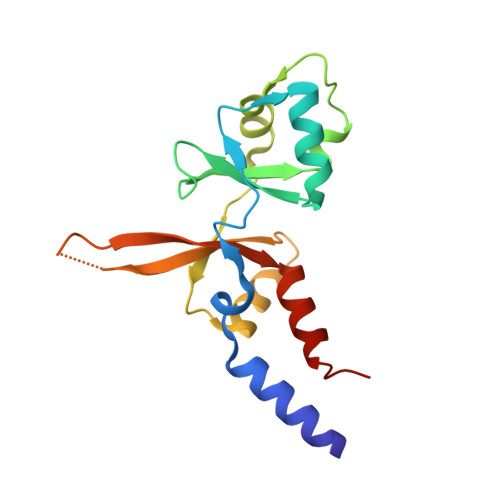
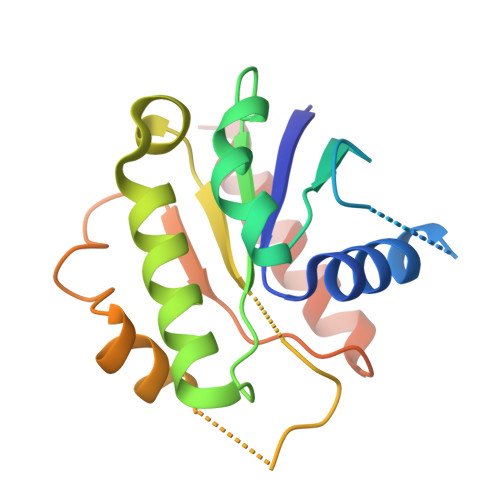
Structural insights into regulation of CNNM-TRPM7 divalent cation uptake by the small GTPase ARL15.
Mahbub, L., Kozlov, G., Zong, P., Lee, E.L., Tetteh, S., Nethramangalath, T., Knorn, C., Jiang, J., Shahsavan, A., Yue, L., Runnels, L., Gehring, K.(2023) Elife 12
- PubMed: 37449820
- DOI: https://doi.org/10.7554/eLife.86129
- Primary Citation of Related Structures:
8F6D - PubMed Abstract:
Cystathionine-β-synthase (CBS)-pair domain divalent metal cation transport mediators (CNNMs) are an evolutionarily conserved family of magnesium transporters. They promote efflux of Mg 2+ ions on their own and influx of divalent cations when expressed with the transient receptor potential ion channel subfamily M member 7 (TRPM7). Recently, ADP-ribosylation factor-like GTPase 15 (ARL15) has been identified as CNNM-binding partner and an inhibitor of divalent cation influx by TRPM7. Here, we characterize ARL15 as a GTP and CNNM-binding protein and demonstrate that ARL15 also inhibits CNNM2 Mg 2+ efflux. The crystal structure of a complex between ARL15 and CNNM2 CBS-pair domain reveals the molecular basis for binding and allowed the identification of mutations that specifically block binding. A binding deficient ARL15 mutant, R95A, failed to inhibit CNNM and TRPM7 transport of Mg 2+ and Zn 2+ ions. Structural analysis and binding experiments with phosphatase of regenerating liver 2 (PRL2 or PTP4A2) showed that ARL15 and PRLs compete for binding CNNM to coordinate regulation of ion transport by CNNM and TRPM7.
- Department of Biochemistry, McGill University, Montreal, Canada.
Organizational Affiliation: