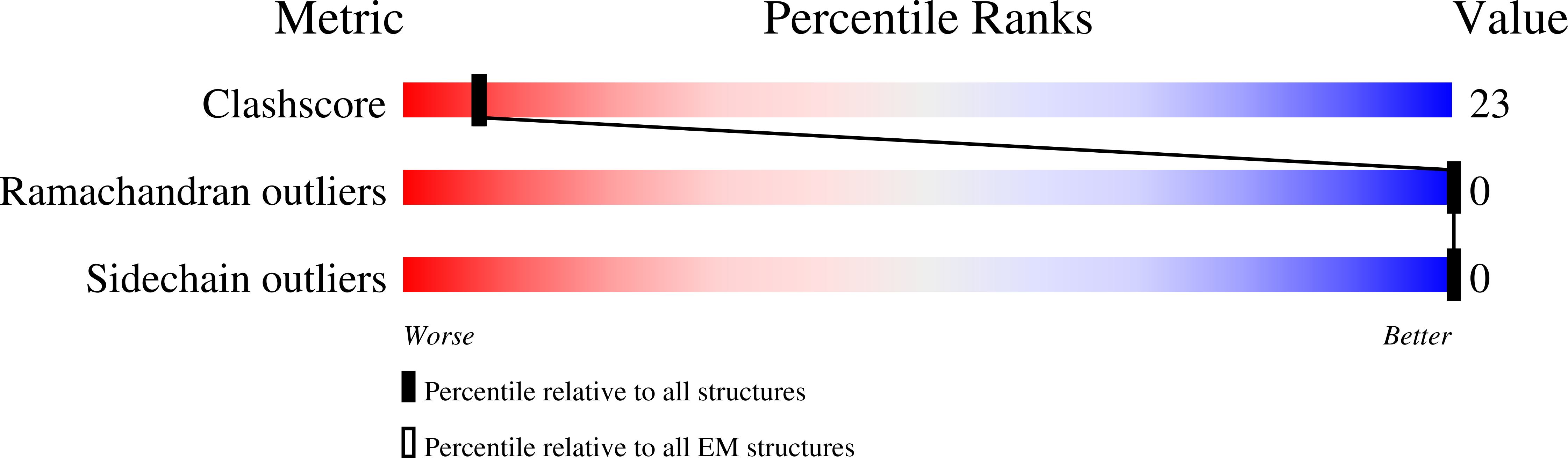Structural basis for GSDMB pore formation and its targeting by IpaH7.8.
Wang, C., Shivcharan, S., Tian, T., Wright, S., Ma, D., Chang, J., Li, K., Song, K., Xu, C., Rathinam, V.A., Ruan, J.(2023) Nature 616: 590-597
- PubMed: 36991122
- DOI: https://doi.org/10.1038/s41586-023-05832-z
- Primary Citation of Related Structures:
8EFP, 8ET1, 8ET2 - PubMed Abstract:
Gasdermins (GSDMs) are pore-forming proteins that play critical roles in host defence through pyroptosis 1,2 . Among GSDMs, GSDMB is unique owing to its distinct lipid-binding profile and a lack of consensus on its pyroptotic potential 3-7 . Recently, GSDMB was shown to exhibit direct bactericidal activity through its pore-forming activity 4 . Shigella, an intracellular, human-adapted enteropathogen, evades this GSDMB-mediated host defence by secreting IpaH7.8, a virulence effector that triggers ubiquitination-dependent proteasomal degradation of GSDMB 4 . Here, we report the cryogenic electron microscopy structures of human GSDMB in complex with Shigella IpaH7.8 and the GSDMB pore. The structure of the GSDMB-IpaH7.8 complex identifies a motif of three negatively charged residues in GSDMB as the structural determinant recognized by IpaH7.8. Human, but not mouse, GSDMD contains this conserved motif, explaining the species specificity of IpaH7.8. The GSDMB pore structure shows the alternative splicing-regulated interdomain linker in GSDMB as a regulator of GSDMB pore formation. GSDMB isoforms with a canonical interdomain linker exhibit normal pyroptotic activity whereas other isoforms exhibit attenuated or no pyroptotic activity. Overall, this work sheds light on the molecular mechanisms of Shigella IpaH7.8 recognition and targeting of GSDMs and shows a structural determinant in GSDMB critical for its pyroptotic activity.
- Department of Immunology, School of Medicine, University of Connecticut Health Center, Farmington, CT, USA.
Organizational Affiliation: