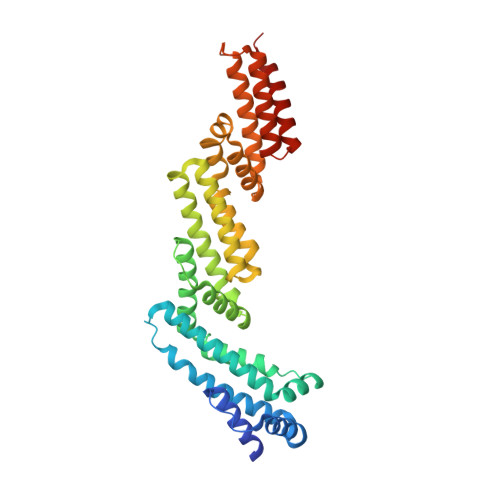
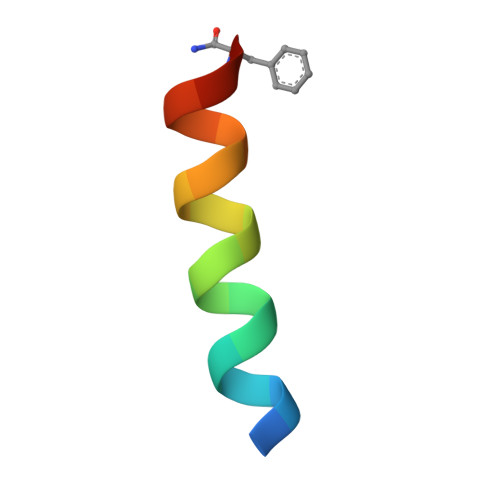
Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Tokareva, O.S., Li, K., Travaline, T.L., Thomson, T.M., Swiecicki, J.M., Moussa, M., Ramirez, J.D., Litchman, S., Verdine, G.L., McGee, J.H.(2023) Nat Commun 14: 6992-6992
- PubMed: 37914719
- DOI: https://doi.org/10.1038/s41467-023-42395-z
- Primary Citation of Related Structures:
8EHZ, 8EI0, 8EI1, 8EI2, 8EI3, 8EI4, 8EI5, 8EI6, 8EI7, 8EI8, 8EI9, 8EIA, 8EIB, 8EIC - PubMed Abstract:
Molecules that induce novel interactions between proteins hold great promise for the study of biological systems and the development of therapeutics, but their discovery has been limited by the complexities of rationally designing interactions between three components, and because known binders to each protein are typically required to inform initial designs. Here, we report a general and rapid method for discovering α-helically constrained (Helicon) polypeptides that cooperatively induce the interaction between two target proteins without relying on previously known binders or an intrinsic affinity between the proteins. We show that Helicons are capable of binding every major class of E3 ubiquitin ligases, which are of great biological and therapeutic interest but remain largely intractable to targeting by small molecules. We then describe a phage-based screening method for discovering "trimerizer" Helicons, and apply it to reprogram E3s to cooperatively bind an enzyme (PPIA), a transcription factor (TEAD4), and a transcriptional coactivator (β-catenin).
- FOG Pharmaceuticals Inc., Cambridge, MA, USA.
Organizational Affiliation: