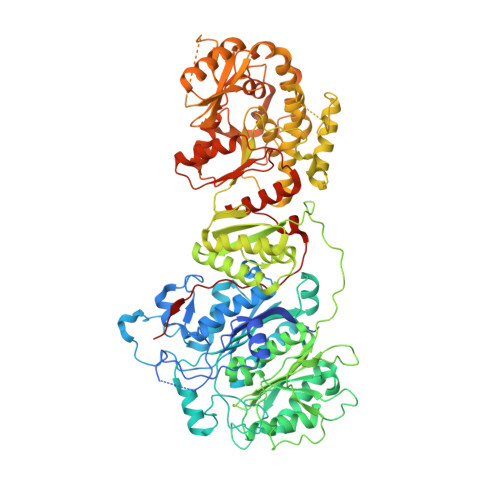
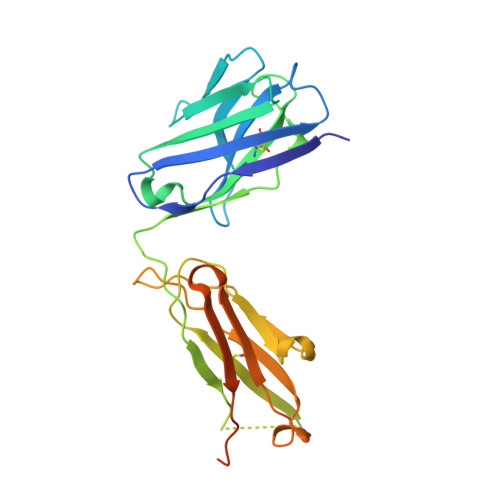
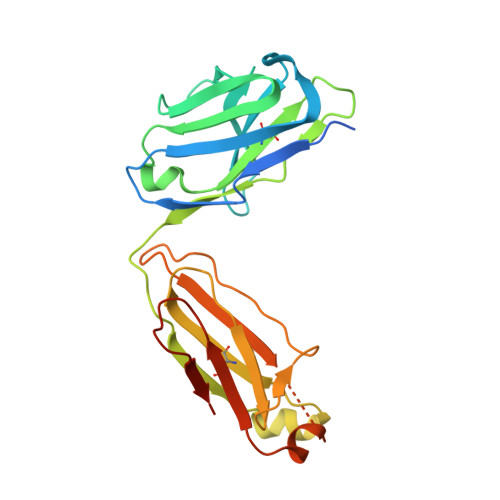
Discovery and Characterization of Antibody Probes of Module 2 of the 6-Deoxyerythronolide B Synthase.
Guzman, K.M., Cogan, D.P., Brodsky, K.L., Soohoo, A.M., Li, X., Sevillano, N., Mathews, I.I., Nguyen, K.P., Craik, C.S., Khosla, C.(2023) Biochemistry 62: 1589-1593
- PubMed: 37184546
- DOI: https://doi.org/10.1021/acs.biochem.3c00156
- Primary Citation of Related Structures:
8EE0, 8EE1 - PubMed Abstract:
Fragment antigen-binding domains of antibodies (F ab s) are powerful probes of structure-function relationships of assembly line polyketide synthases (PKSs). We report the discovery and characterization of F ab s interrogating the structure and function of the ketosynthase-acyltransferase (KS-AT) core of Module 2 of the 6-deoxyerythronolide B synthase (DEBS). Two F ab s (AC2 and BB1) were identified to potently inhibit the catalytic activity of Module 2. Both AC2 and BB1 were found to modulate ACP-mediated reactions catalyzed by this module, albeit by distinct mechanisms. AC2 primarily affects the rate ( k cat ), whereas BB1 increases the K M of an ACP-mediated reaction. A third F ab , AA5, binds to the KS-AT fragment of DEBS Module 2 without altering either parameter; it is phenotypically reminiscent of a previously characterized F ab , 1B2, shown to principally recognize the N-terminal helical docking domain of DEBS Module 3. Crystal structures of AA5 and 1B2 bound to the KS-AT fragment of Module 2 were solved to 2.70 and 2.65 Å resolution, respectively, and revealed entirely distinct recognition features of the two antibodies. The new tools and insights reported here pave the way toward advancing our understanding of the structure-function relationships of DEBS Module 2, arguably the most well-studied module of an assembly line PKS.
- Department of Chemical Engineering, Stanford University, Stanford, California 94305, United States.
Organizational Affiliation: