Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Kumar, S., Zhou, Y.(2023) Nat Commun
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | 946 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: SPY, At3g11540, F24K9.29 EC: 2.4.1.255 (PDB Primary Data), 2.4.1.221 (PDB Primary Data) | 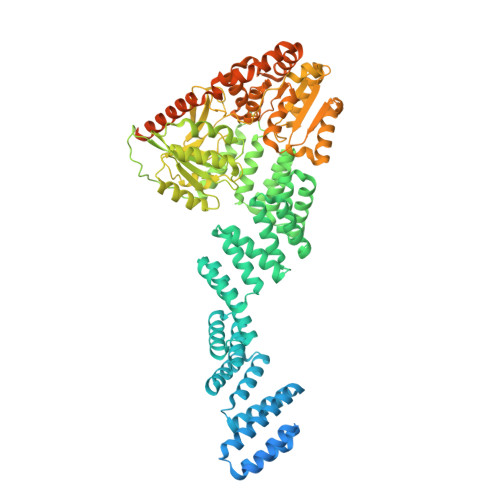 | |
UniProt | |||||
Find proteins for Q96301 (Arabidopsis thaliana) Explore Q96301 Go to UniProtKB: Q96301 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96301 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | -- |