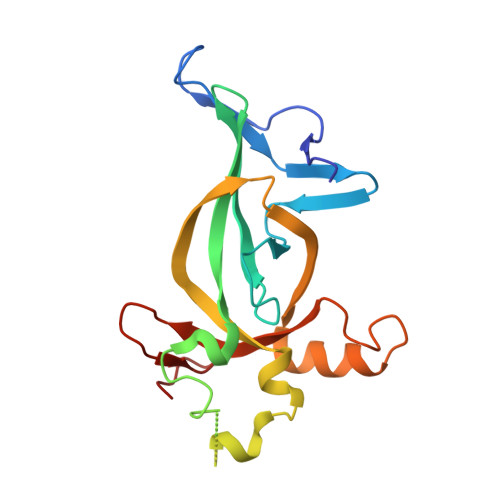
TNRC18 engages H3K9me3 to mediate silencing of endogenous retrotransposons.
Zhao, S., Lu, J., Pan, B., Fan, H., Byrum, S.D., Xu, C., Kim, A., Guo, Y., Kanchi, K.L., Gong, W., Sun, T., Storey, A.J., Burkholder, N.T., Mackintosh, S.G., Kuhlers, P.C., Edmondson, R.D., Strahl, B.D., Diao, Y., Tackett, A.J., Raab, J.R., Cai, L., Song, J., Wang, G.G.(2023) Nature 623: 633-642
- PubMed: 37938770
- DOI: https://doi.org/10.1038/s41586-023-06688-z
- Primary Citation of Related Structures:
8DS8 - PubMed Abstract:
Trimethylation of histone H3 lysine 9 (H3K9me3) is crucial for the regulation of gene repression and heterochromatin formation, cell-fate determination and organismal development 1 . H3K9me3 also provides an essential mechanism for silencing transposable elements 1-4 . However, previous studies have shown that canonical H3K9me3 readers (for example, HP1 (refs. 5-9 ) and MPP8 (refs. 10-12 )) have limited roles in silencing endogenous retroviruses (ERVs), one of the main transposable element classes in the mammalian genome 13 . Here we report that trinucleotide-repeat-containing 18 (TNRC18), a poorly understood chromatin regulator, recognizes H3K9me3 to mediate the silencing of ERV class I (ERV1) elements such as LTR12 (ref. 14 ). Biochemical, biophysical and structural studies identified the carboxy-terminal bromo-adjacent homology (BAH) domain of TNRC18 (TNRC18(BAH)) as an H3K9me3-specific reader. Moreover, the amino-terminal segment of TNRC18 is a platform for the direct recruitment of co-repressors such as HDAC-Sin3-NCoR complexes, thus enforcing optimal repression of the H3K9me3-demarcated ERVs. Point mutagenesis that disrupts the TNRC18(BAH)-mediated H3K9me3 engagement caused neonatal death in mice and, in multiple mammalian cell models, led to derepressed expression of ERVs, which affected the landscape of cis-regulatory elements and, therefore, gene-expression programmes. Collectively, we describe a new H3K9me3-sensing and regulatory pathway that operates to epigenetically silence evolutionarily young ERVs and exert substantial effects on host genome integrity, transcriptomic regulation, immunity and development.
- Department of Pharmacology and Cancer Biology, Duke University School of Medicine, Durham, NC, USA.
Organizational Affiliation: