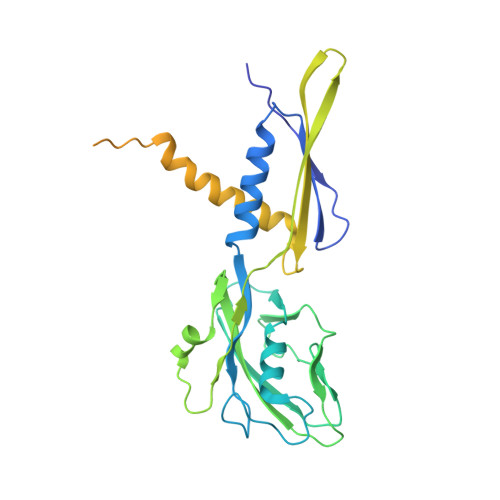
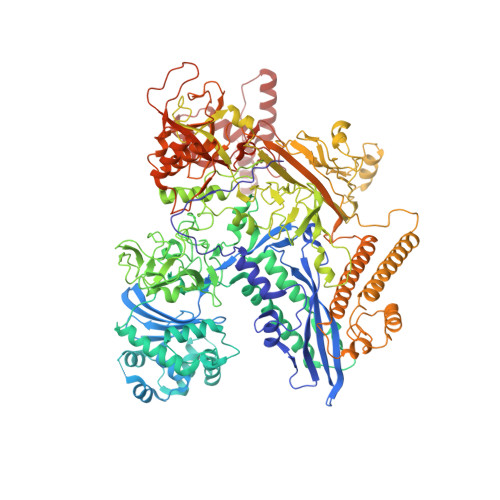
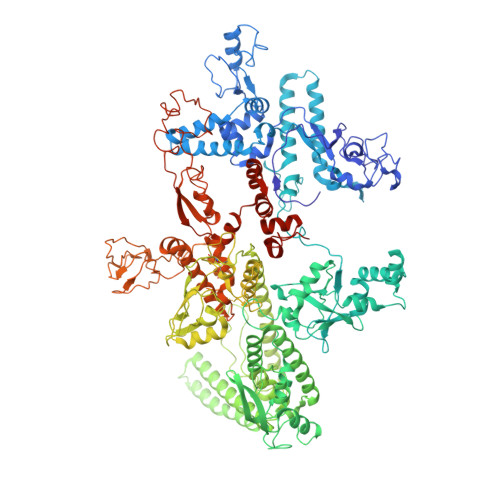
Cryo-EM Structure of Porphyromonas gingivalis RNA Polymerase.
Bu, F., Wang, X., Li, M., Ma, L., Wang, C., Hu, Y., Cao, Z., Liu, B.(2024) J Mol Biology 436: 168568-168568
- PubMed: 38583515
- DOI: https://doi.org/10.1016/j.jmb.2024.168568
- Primary Citation of Related Structures:
8DKC - PubMed Abstract:
Porphyromonas gingivalis, an anaerobic CFB (Cytophaga, Fusobacterium, and Bacteroides) group bacterium, is the keystone pathogen of periodontitis and has been implicated in various systemic diseases. Increased antibiotic resistance and lack of effective antibiotics necessitate a search for new intervention strategies. Here we report a 3.5 Å resolution cryo-EM structure of P. gingivalis RNA polymerase (RNAP). The structure displays new structural features in its ω subunit and multiple domains in β and β' subunits, which differ from their counterparts in other bacterial RNAPs. Superimpositions with E. coli RNAP holoenzyme and initiation complex further suggest that its ω subunit may contact the σ4 domain, thereby possibly contributing to the assembly and stabilization of initiation complexes. In addition to revealing the unique features of P. gingivalis RNAP, our work offers a framework for future studies of transcription regulation in this important pathogen, as well as for structure-based drug development.
- Section of Transcription & Gene Regulation, The Hormel Institute, University of Minnesota, Austin, MN, USA.
Organizational Affiliation: