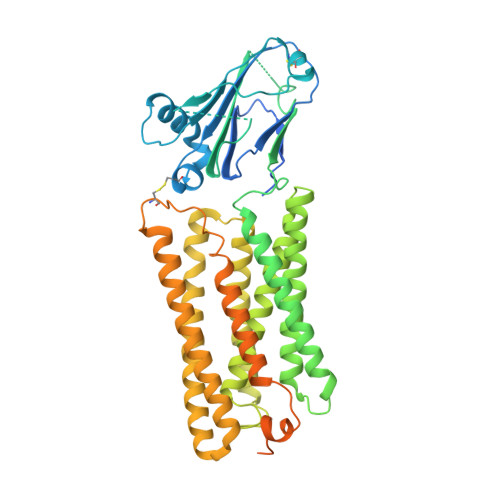
Structure of the GOLD-domain seven-transmembrane helix protein family member TMEM87A.
Hoel, C.M., Zhang, L., Brohawn, S.G.(2022) Elife 11
- PubMed: 36373655
- DOI: https://doi.org/10.7554/eLife.81704
- Primary Citation of Related Structures:
8CTJ - PubMed Abstract:
TMEM87s are eukaryotic transmembrane proteins with two members (TMEM87A and TMEM87B) in humans. TMEM87s have proposed roles in protein transport to and from the Golgi, as mechanosensitive ion channels, and in developmental signaling. TMEM87 disruption has been implicated in cancers and developmental disorders. To better understand TMEM87 structure and function, we determined a cryo-EM structure of human TMEM87A in lipid nanodiscs. TMEM87A consists of a Golgi-dynamics (GOLD) domain atop a membrane-spanning seven-transmembrane helix domain with a large cavity open to solution and the membrane outer leaflet. Structural and functional analyses suggest TMEM87A may not function as an ion channel or G-protein coupled receptor. We find TMEM87A shares its characteristic domain arrangement with seven other proteins in humans; three that had been identified as evolutionary related (TMEM87B, GPR107, and GPR108) and four previously unrecognized homologs (GPR180, TMEM145, TMEM181, and WLS). Among these structurally related GO LD domain s even- t ransmembrane helix (GOST) proteins, WLS is best characterized as a membrane trafficking and secretion chaperone for lipidated Wnt signaling proteins. We find key structural determinants for WLS function are conserved in TMEM87A. We propose TMEM87A and structurally homologous GOST proteins could serve a common role in trafficking membrane-associated cargo.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, United States.
Organizational Affiliation: