Molecular Analysis of pSK1 par: A Novel Plasmid Partitioning System Encoded by Staphylococcal Multiresistance Plasmids.
Chan, H.Y., Jensen, S.O., LeBard, R.J., Figgett, W.A., Lai, E., Simpson, A.E., Brzoska, A.J., Davies, D.S., Connolly, A.M., Cordwell, S.J., Travis, B.A., Salinas, R., Skurray, R.A., Firth, N., Schumacher, M.A.(2022) J Mol Biology 434: 167770-167770
- PubMed: 35907571
- DOI: https://doi.org/10.1016/j.jmb.2022.167770
- Primary Citation of Related Structures:
8CSH - PubMed Abstract:
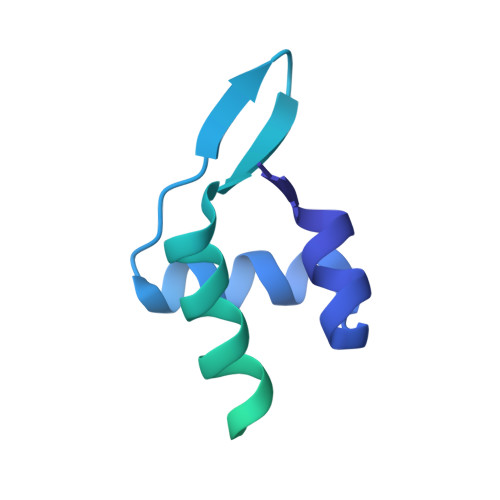
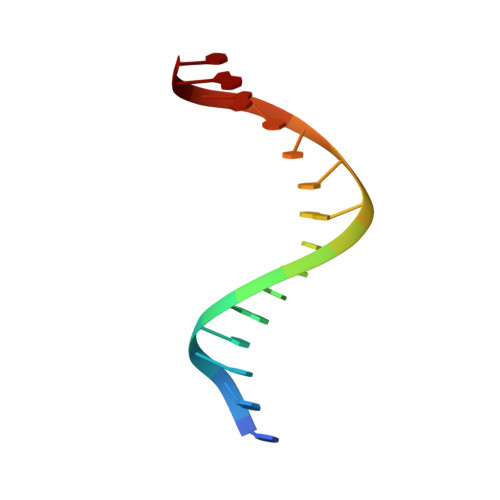
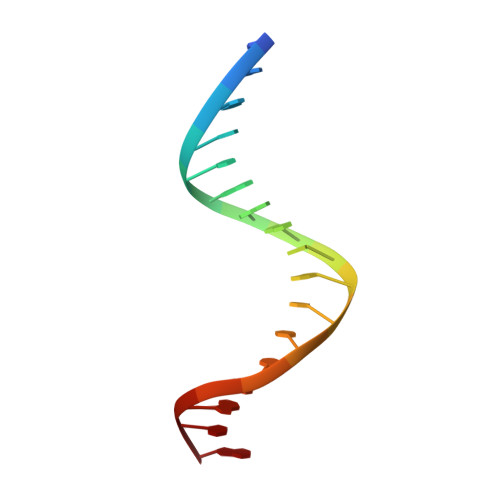
The segregation of prokaryotic plasmids typically requires a centromere-like site and two proteins, a centromere-binding protein (CBP) and an NTPase. By contrast, a single 245 residue Par protein mediates partition of the prototypical staphylococcal multiresistance plasmid pSK1 in the absence of an identifiable NTPase component. To gain insight into centromere binding by pSK1 Par and its segregation function we performed structural, biochemical and in vivo studies. Here we show that pSK1 Par binds a centromere consisting of seven repeat elements. We demonstrate this Par-centromere interaction also mediates Par autoregulation. To elucidate the Par centromere binding mechanism, we obtained a structure of the Par N-terminal DNA-binding domain bound to centromere DNA to 2.25 Å. The pSK1 Par structure, which harbors a winged-helix-turn-helix (wHTH), is distinct from other plasmid CBP structures but shows homology to the B. subtilis chromosome segregation protein, RacA. Biochemical studies suggest the region C-terminal to the Par wHTH forms coiled coils and mediates oligomerization. Fluorescence microscopy analyses show that pSK1 Par enhances the separation of plasmids from clusters, driving effective segregation upon cell division. Combined the data provide insight into the molecular properties of a single protein partition system.
- School of Life and Environmental Sciences, University of Sydney, New South Wales 2006, Australia.
Organizational Affiliation: