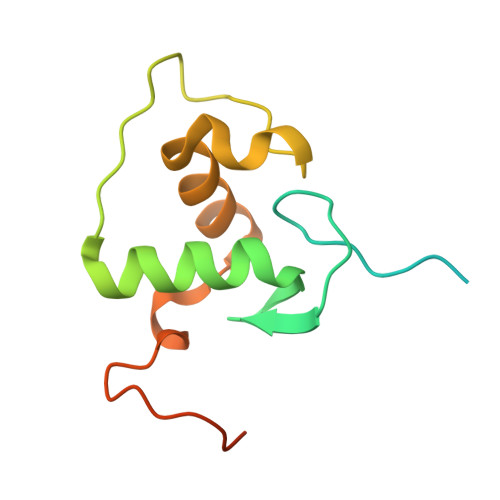
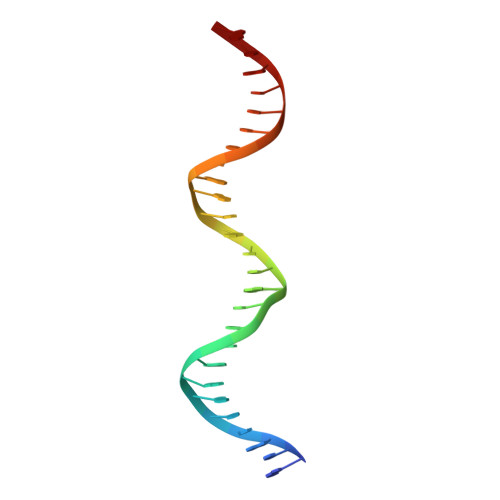
Asymmetric dimerization in a transcription factor superfamily is promoted by allosteric interactions with DNA.
Patel, A.K.M., Vilela, P., Shaik, T.B., McEwen, A.G., Hazemann, I., Brillet, K., Ennifar, E., Hamiche, A., Markov, G.V., Laudet, V., Moras, D., Klaholz, B.P., Billas, I.M.L.(2023) Nucleic Acids Res 51: 8864-8879
- PubMed: 37503845
- DOI: https://doi.org/10.1093/nar/gkad632
- Primary Citation of Related Structures:
8CEF - PubMed Abstract:
Transcription factors, such as nuclear receptors achieve precise transcriptional regulation by means of a tight and reciprocal communication with DNA, where cooperativity gained by receptor dimerization is added to binding site sequence specificity to expand the range of DNA target gene sequences. To unravel the evolutionary steps in the emergence of DNA selection by steroid receptors (SRs) from monomeric to dimeric palindromic binding sites, we carried out crystallographic, biophysical and phylogenetic studies, focusing on the estrogen-related receptors (ERRs, NR3B) that represent closest relatives of SRs. Our results, showing the structure of the ERR DNA-binding domain bound to a palindromic response element (RE), unveil the molecular mechanisms of ERR dimerization which are imprinted in the protein itself with DNA acting as an allosteric driver by allowing the formation of a novel extended asymmetric dimerization region (KR-box). Phylogenetic analyses suggest that this dimerization asymmetry is an ancestral feature necessary for establishing a strong overall dimerization interface, which was progressively modified in other SRs in the course of evolution.
- IGBMC (Institute of Genetics and of Molecular and Cellular Biology), Centre for Integrative Biology (CBI), Illkirch, France.
Organizational Affiliation: