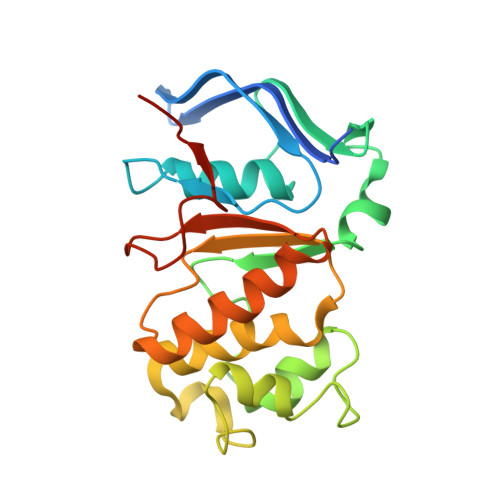
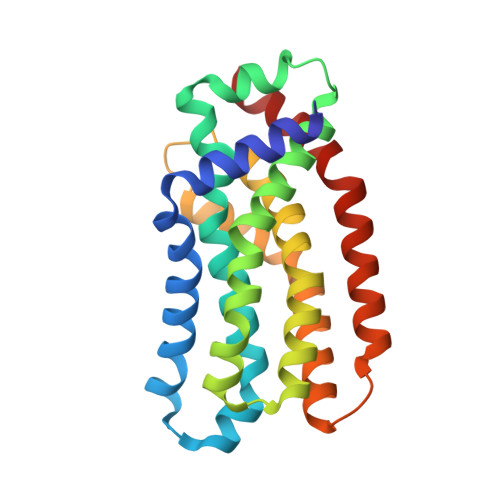
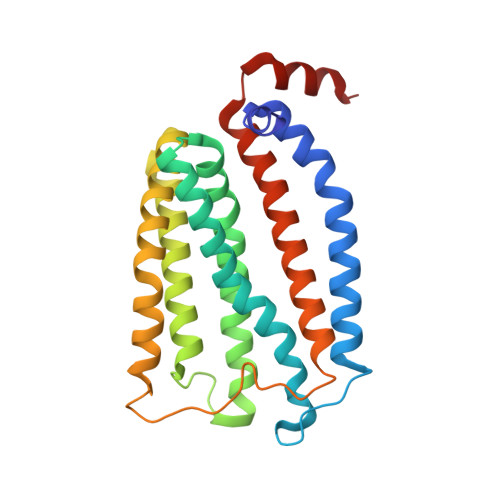
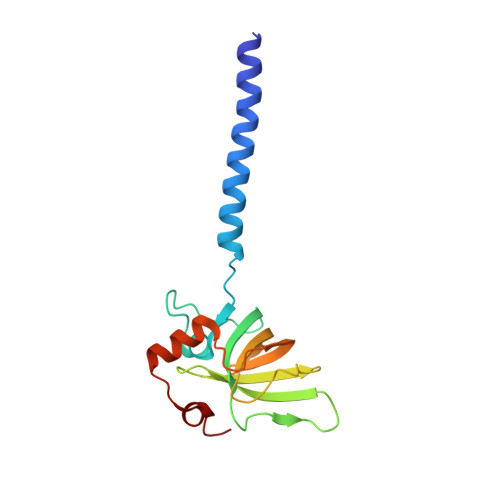
Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Ilcu, L., Denkhaus, L., Brausemann, A., Zhang, L., Einsle, O.(2023) Nat Commun 14: 5190-5190
- PubMed: 37626034
- DOI: https://doi.org/10.1038/s41467-023-40881-y
- Primary Citation of Related Structures:
8CE1, 8CE5, 8CE8, 8CEA - PubMed Abstract:
Mono- and multiheme cytochromes c are post-translationally matured by the covalent attachment of heme. For this, Escherichia coli employs the most complex type of maturation machineries, the Ccm-system (for cytochrome c maturation). It consists of two membrane protein complexes, one of which shuttles heme across the membrane to a mobile chaperone that then delivers the cofactor to the second complex, an apoprotein:heme lyase, for covalent attachment. Here we report cryo-electron microscopic structures of the heme translocation complex CcmABCD from E. coli, alone and bound to the heme chaperone CcmE. CcmABCD forms a heterooctameric complex centered around the ABC transporter CcmAB that does not by itself transport heme. Our data suggest that the complex flops a heme group from the inner to the outer leaflet at its CcmBC interfaces, driven by ATP hydrolysis at CcmA. A conserved heme-handling motif (WxWD) at the periplasmic side of CcmC rotates the heme by 90° for covalent attachment to the heme chaperone CcmE that we find interacting exclusively with the CcmB subunit.
- Institut für Biochemie, Albert-Ludwigs-Universität Freiburg, 79104, Freiburg im Breisgau, Germany.
Organizational Affiliation: