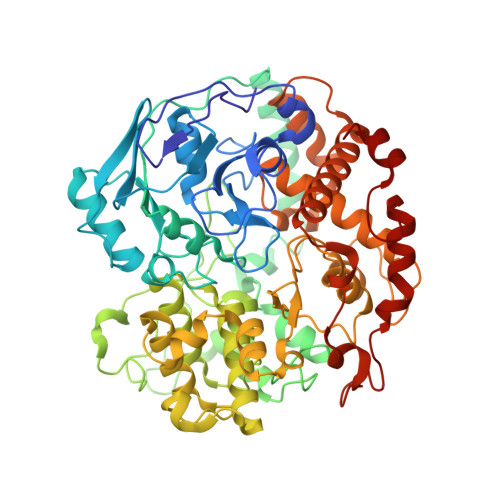
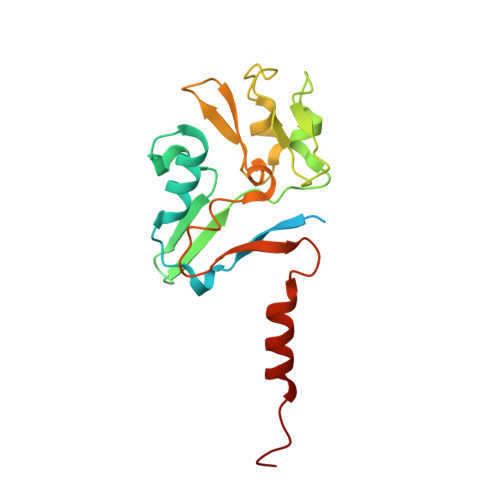
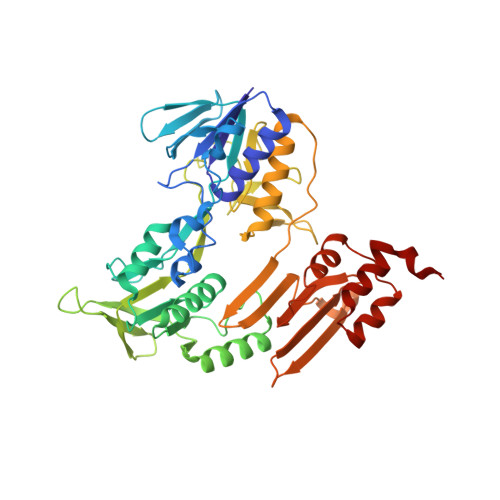
A bacterial tungsten-containing aldehyde oxidoreductase forms an enzymatic decorated protein nanowire.
Winiarska, A., Ramirez-Amador, F., Hege, D., Gemmecker, Y., Prinz, S., Hochberg, G., Heider, J., Szaleniec, M., Schuller, J.M.(2023) Sci Adv 9: eadg6689-eadg6689
- PubMed: 37267359
- DOI: https://doi.org/10.1126/sciadv.adg6689
- Primary Citation of Related Structures:
8C0Z - PubMed Abstract:
Aldehyde oxidoreductases (AORs) are tungsten enzymes catalyzing the oxidation of many different aldehydes to the corresponding carboxylic acids. In contrast to other known AORs, the enzyme from the denitrifying betaproteobacterium Aromatoleum aromaticum (AOR Aa ) consists of three different subunits (AorABC) and uses nicotinamide adenine dinucleotide (NAD) as an electron acceptor. Here, we reveal that the enzyme forms filaments of repeating AorAB protomers that are capped by a single NAD-binding AorC subunit, based on solving its structure via cryo-electron microscopy. The polyferredoxin-like subunit AorA oligomerizes to an electron-conducting nanowire that is decorated with enzymatically active and W-cofactor (W-co) containing AorB subunits. Our structure further reveals the binding mode of the native substrate benzoate in the AorB active site. This, together with quantum mechanics:molecular mechanics (QM:MM)-based modeling for the coordination of the W-co, enables formulation of a hypothetical catalytic mechanism that paves the way to further engineering for applications in synthetic biology and biotechnology.
- Jerzy Haber Institute of Catalysis and Surface Chemistry Polish Academy of Sciences, Kraków, Poland.
Organizational Affiliation: