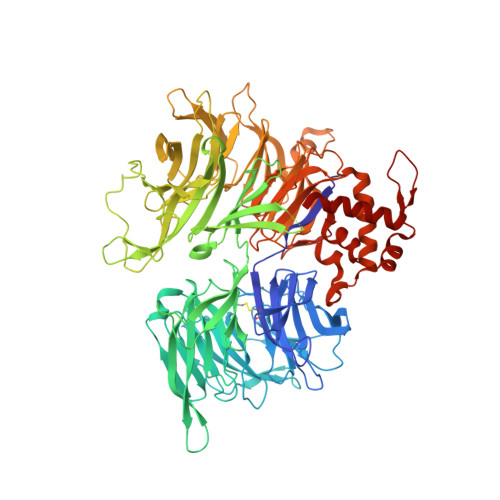
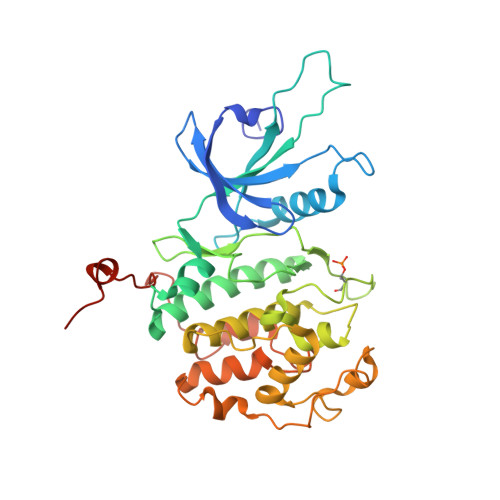
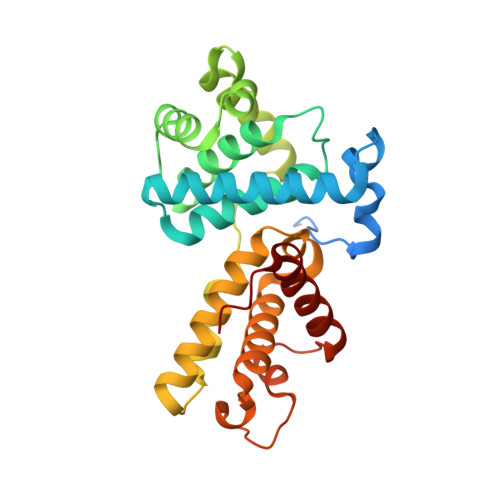
Design principles for cyclin K molecular glue degraders.
Kozicka, Z., Suchyta, D.J., Focht, V., Kempf, G., Petzold, G., Jentzsch, M., Zou, C., Di Genua, C., Donovan, K.A., Coomar, S., Cigler, M., Mayor-Ruiz, C., Schmid-Burgk, J.L., Haussinger, D., Winter, G.E., Fischer, E.S., Slabicki, M., Gillingham, D., Ebert, B.L., Thoma, N.H.(2024) Nat Chem Biol 20: 93-102
- PubMed: 37679459
- DOI: https://doi.org/10.1038/s41589-023-01409-z
- Primary Citation of Related Structures:
8BU1, 8BU2, 8BU3, 8BU4, 8BU5, 8BU6, 8BU7, 8BU9, 8BUA, 8BUB, 8BUC, 8BUD, 8BUE, 8BUF, 8BUG, 8BUH, 8BUI, 8BUJ, 8BUK, 8BUL, 8BUM, 8BUN, 8BUO, 8BUP, 8BUQ, 8BUR, 8BUS, 8BUT - PubMed Abstract:
Molecular glue degraders are an effective therapeutic modality, but their design principles are not well understood. Recently, several unexpectedly diverse compounds were reported to deplete cyclin K by linking CDK12-cyclin K to the DDB1-CUL4-RBX1 E3 ligase. Here, to investigate how chemically dissimilar small molecules trigger cyclin K degradation, we evaluated 91 candidate degraders in structural, biophysical and cellular studies and reveal all compounds acquire glue activity via simultaneous CDK12 binding and engagement of DDB1 interfacial residues, in particular Arg928. While we identify multiple published kinase inhibitors as cryptic degraders, we also show that these glues do not require pronounced inhibitory properties for activity and that the relative degree of CDK12 inhibition versus cyclin K degradation is tuneable. We further demonstrate cyclin K degraders have transcriptional signatures distinct from CDK12 inhibitors, thereby offering unique therapeutic opportunities. The systematic structure-activity relationship analysis presented herein provides a conceptual framework for rational molecular glue design.
- Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: