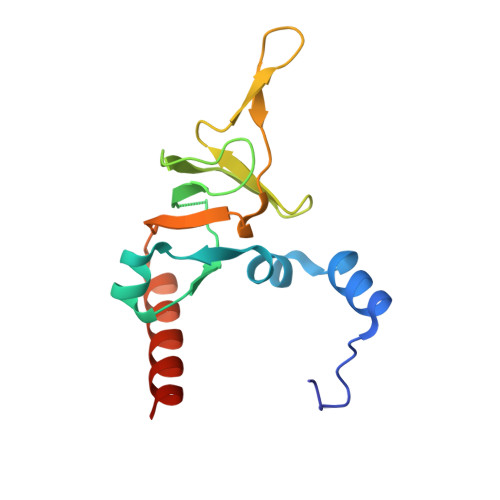
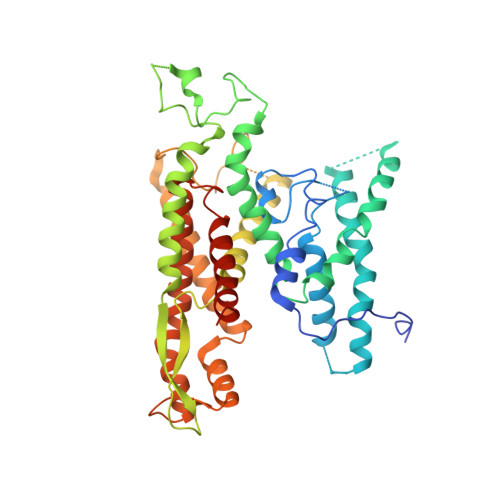
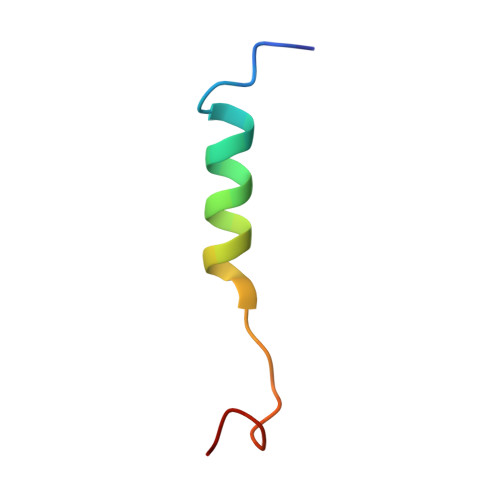
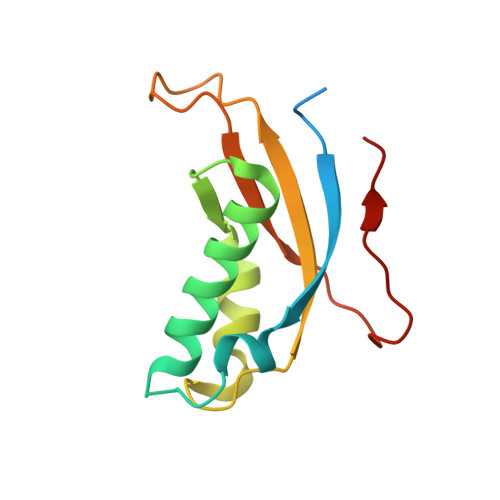
Molecular view of ER membrane remodeling by the Sec61/TRAP translocon.
Karki, S., Javanainen, M., Rehan, S., Tranter, D., Kellosalo, J., Huiskonen, J.T., Happonen, L., Paavilainen, V.(2023) EMBO Rep 24: e57910-e57910
- PubMed: 37983950
- DOI: https://doi.org/10.15252/embr.202357910
- Primary Citation of Related Structures:
8BF9 - PubMed Abstract:
Protein translocation across the endoplasmic reticulum (ER) membrane is an essential step during protein entry into the secretory pathway. The conserved Sec61 protein-conducting channel facilitates polypeptide translocation and coordinates cotranslational polypeptide-processing events. In cells, the majority of Sec61 is stably associated with a heterotetrameric membrane protein complex, the translocon-associated protein complex (TRAP), yet the mechanism by which TRAP assists in polypeptide translocation remains unknown. Here, we present the structure of the core Sec61/TRAP complex bound to a mammalian ribosome by cryogenic electron microscopy (cryo-EM). Ribosome interactions anchor the Sec61/TRAP complex in a conformation that renders the ER membrane locally thinner by significantly curving its lumenal leaflet. We propose that TRAP stabilizes the ribosome exit tunnel to assist nascent polypeptide insertion through Sec61 and provides a ratcheting mechanism into the ER lumen mediated by direct polypeptide interactions.
- Institute of Biotechnology, University of Helsinki, Helsinki, Finland.
Organizational Affiliation: