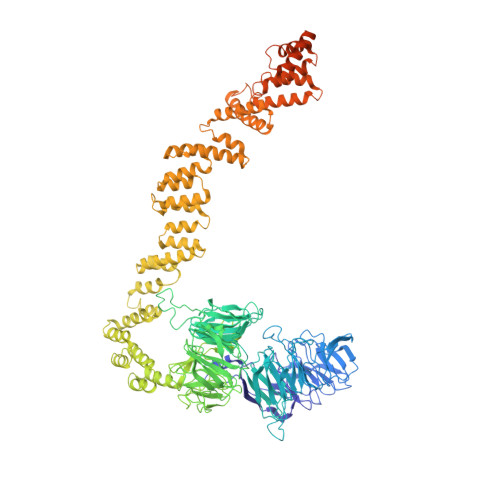
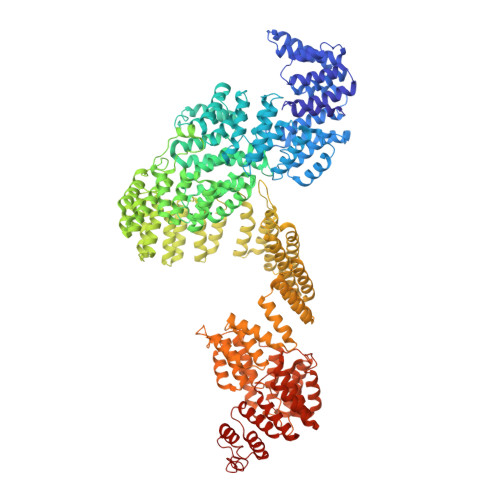
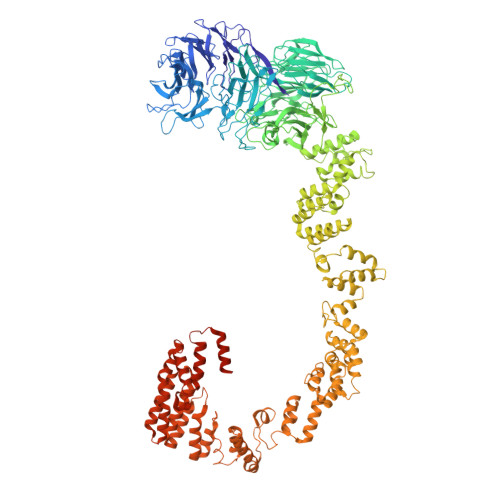
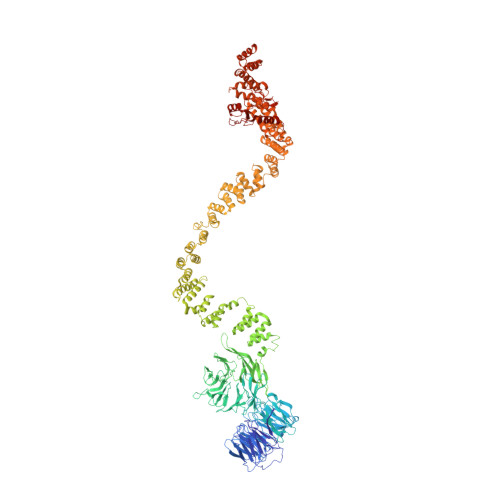
The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Lacey, S.E., Foster, H.E., Pigino, G.(2023) Nat Struct Mol Biol 30: 584-593
- PubMed: 36593313
- DOI: https://doi.org/10.1038/s41594-022-00905-5
- Primary Citation of Related Structures:
8BD7, 8BDA - PubMed Abstract:
Anterograde intraflagellar transport (IFT) trains are essential for cilia assembly and maintenance. These trains are formed of 22 IFT-A and IFT-B proteins that link structural and signaling cargos to microtubule motors for import into cilia. It remains unknown how the IFT-A/-B proteins are arranged into complexes and how these complexes polymerize into functional trains. Here we use in situ cryo-electron tomography of Chlamydomonas reinhardtii cilia and AlphaFold2 protein structure predictions to generate a molecular model of the entire anterograde train. We show how the conformations of both IFT-A and IFT-B are dependent on lateral interactions with neighboring repeats, suggesting that polymerization is required to cooperatively stabilize the complexes. Following three-dimensional classification, we reveal how IFT-B extends two flexible tethers to maintain a connection with IFT-A that can withstand the mechanical stresses present in actively beating cilia. Overall, our findings provide a framework for understanding the fundamental processes that govern cilia assembly.
- Human Technopole, Milan, Italy.
Organizational Affiliation: