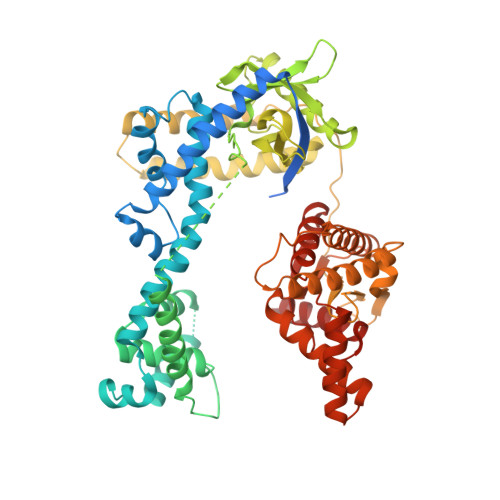
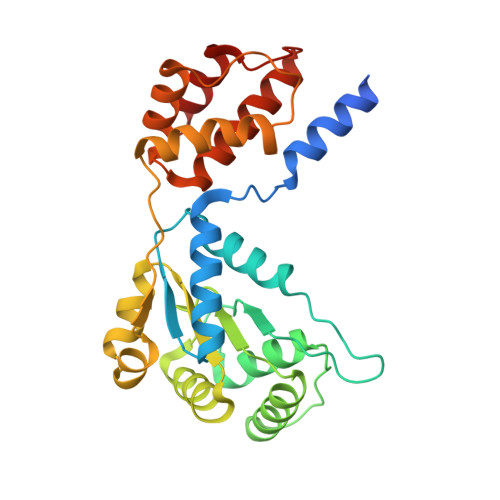
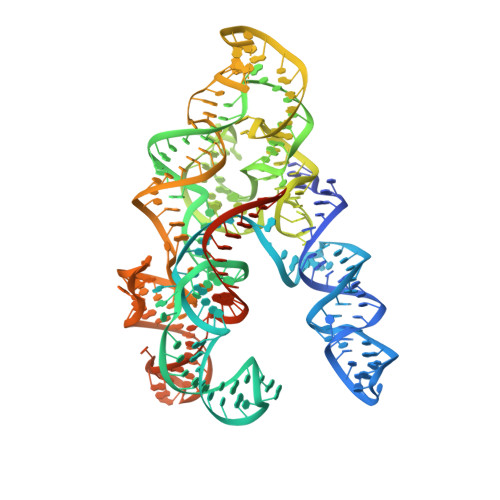
Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Schmitz, M., Querques, I., Oberli, S., Chanez, C., Jinek, M.(2022) Cell 185: 4999
- PubMed: 36435179
- DOI: https://doi.org/10.1016/j.cell.2022.11.009
- Primary Citation of Related Structures:
8BD4, 8BD5, 8BD6 - PubMed Abstract:
CRISPR-Cas systems have been co-opted by Tn7-like transposable elements to direct RNA-guided transposition. Type V-K CRISPR-associated transposons rely on the concerted activities of the pseudonuclease Cas12k, the AAA+ ATPase TnsC, the Zn-finger protein TniQ, and the transposase TnsB. Here we present a cryo-electron microscopic structure of a target DNA-bound Cas12k-transposon recruitment complex comprised of RNA-guided Cas12k, TniQ, a polymeric TnsC filament and, unexpectedly, the ribosomal protein S15. Complex assembly, mediated by a network of interactions involving the guide RNA, TniQ, and S15, results in R-loop completion. TniQ contacts two TnsC protomers at the Cas12k-proximal filament end, likely nucleating its polymerization. Transposition activity assays corroborate our structural findings, implying that S15 is a bona fide component of the type V crRNA-guided transposon machinery. Altogether, our work uncovers key mechanistic aspects underpinning RNA-mediated assembly of CRISPR-associated transposons to guide their development as programmable tools for site-specific insertion of large DNA payloads.
- Department of Biochemistry, University of Zurich, Zurich 8057, Switzerland.
Organizational Affiliation: