Photosystem I assembly intermediate of Avena sativa
Naschberger, A., Amunts, A., Nelson, N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I P700 chlorophyll a apoprotein A1 | 750 | Avena sativa | Mutation(s): 0 EC: 1.97.1.12 Membrane Entity: Yes | 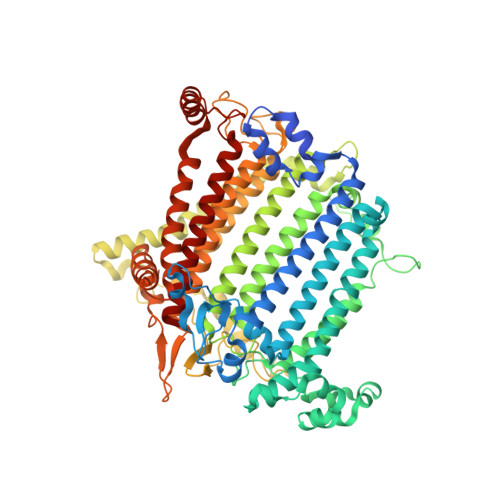 | |
UniProt | |||||
Find proteins for A0A3G1AUL2 (Avena sativa) Explore A0A3G1AUL2 Go to UniProtKB: A0A3G1AUL2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A3G1AUL2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I P700 chlorophyll a apoprotein A2 | 734 | Avena sativa | Mutation(s): 0 EC: 1.97.1.12 Membrane Entity: Yes | 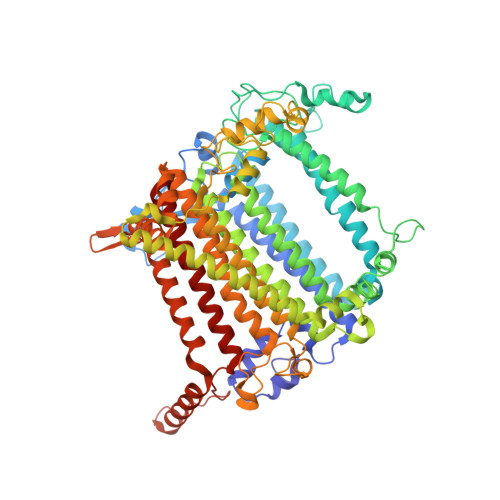 | |
UniProt | |||||
Find proteins for A0A3G1AZK6 (Avena sativa) Explore A0A3G1AZK6 Go to UniProtKB: A0A3G1AZK6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A3G1AZK6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I iron-sulfur center | 81 | Avena sativa | Mutation(s): 0 EC: 1.97.1.12 Membrane Entity: Yes | 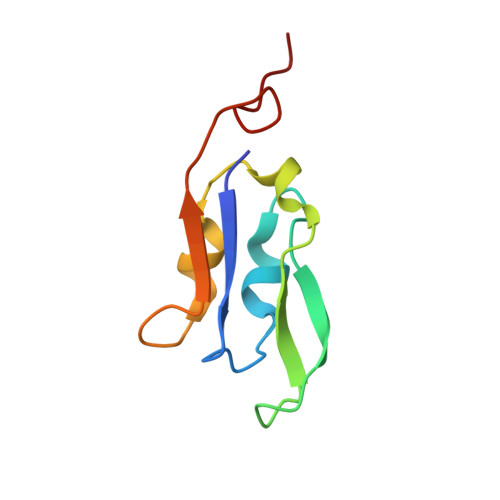 | |
UniProt | |||||
Find proteins for A0A3G1ATU7 (Avena sativa) Explore A0A3G1ATU7 Go to UniProtKB: A0A3G1ATU7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A3G1ATU7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit II, chloroplastic | 206 | Avena sativa | Mutation(s): 0 | 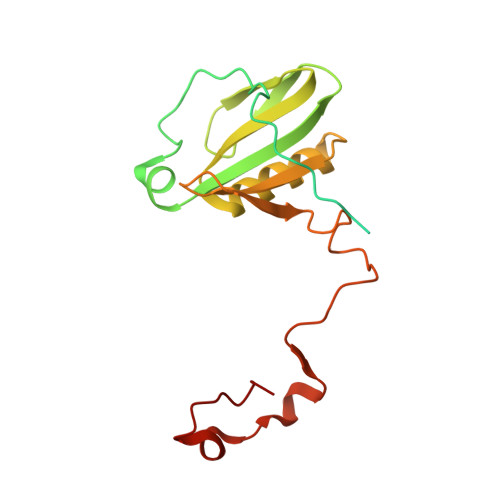 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit IV A, chloroplastic | 143 | Avena sativa | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit VI, chloroplastic | F [auth H] | 94 | Avena sativa | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem I reaction center subunit VIII | G [auth I] | 33 | Avena sativa | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for A0A3G1AZM6 (Avena sativa) Explore A0A3G1AZM6 Go to UniProtKB: A0A3G1AZM6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A3G1AZM6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PSI subunit V | H [auth L] | 213 | Avena sativa | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DGD (Subject of Investigation/LOI) Query on DGD | GD [auth B] | DIGALACTOSYL DIACYL GLYCEROL (DGDG) C51 H96 O15 LDQFLSUQYHBXSX-HXXRYREZSA-N |  | ||
| CLA (Subject of Investigation/LOI) Query on CLA | AA [auth A] AB [auth A] AC [auth B] AD [auth B] BA [auth A] | CHLOROPHYLL A C55 H72 Mg N4 O5 ATNHDLDRLWWWCB-AENOIHSZSA-M |  | ||
| CL0 (Subject of Investigation/LOI) Query on CL0 | I [auth A], KD [auth H] | CHLOROPHYLL A ISOMER C55 H72 Mg N4 O5 VIQFHHZSLDFWDU-DVXFRRMCSA-M |  | ||
| LMG (Subject of Investigation/LOI) Query on LMG | NB [auth A] | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE C45 H86 O10 DCLTVZLYPPIIID-CVELTQQQSA-N |  | ||
| LHG (Subject of Investigation/LOI) Query on LHG | BB [auth A], CB [auth A], HD [auth B] | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE C38 H75 O10 P BIABMEZBCHDPBV-MPQUPPDSSA-N |  | ||
| BCR (Subject of Investigation/LOI) Query on BCR | CD [auth B] DB [auth A] DD [auth B] EB [auth A] ED [auth B] | BETA-CAROTENE C40 H56 OENHQHLEOONYIE-JLTXGRSLSA-N |  | ||
| LMU Query on LMU | KB [auth A], MB [auth A], OB [auth A] | DODECYL-ALPHA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-YHBSTRCHSA-N |  | ||
| PQN (Subject of Investigation/LOI) Query on PQN | BD [auth B], ZA [auth A] | PHYLLOQUINONE C31 H46 O2 MBWXNTAXLNYFJB-NKFFZRIASA-N |  | ||
| SF4 (Subject of Investigation/LOI) Query on SF4 | IB [auth A], ID [auth C], JD [auth C] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| The Swedish Foundation for Strategic Research | Sweden | ARC19-0051 |
| Knut and Alice Wallenberg Foundation | Sweden | 2018.0080 |
| Israel Science Foundation | Israel | 569/17 |