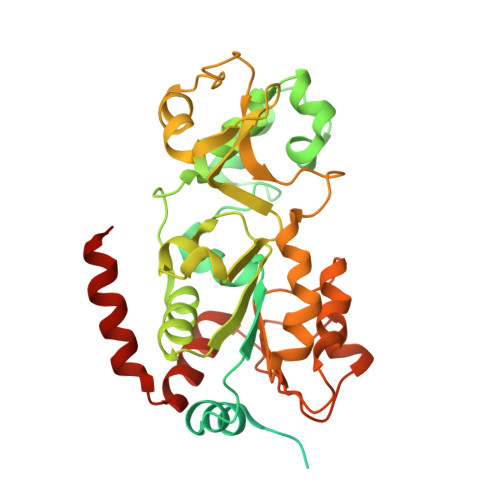
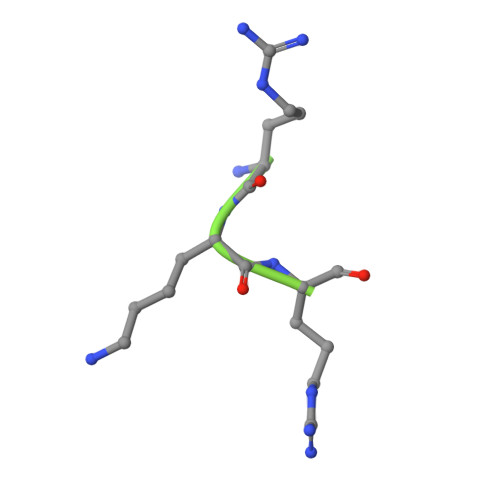
Molecular Mechanism of Sirtuin 1 Modulation by the AROS Protein.
Weiss, S., Adolph, R.S., Schweimer, K., DiFonzo, A., Meleshin, M., Schutkowski, M., Steegborn, C.(2022) Int J Mol Sci 23
- PubMed: 36361557
- DOI: https://doi.org/10.3390/ijms232112764
- Primary Citation of Related Structures:
8BBK - PubMed Abstract:
The protein lysine deacylases of the NAD + -dependent Sirtuin family contribute to metabolic regulation, stress responses, and aging processes, and the human Sirtuin isoforms, Sirt1-7, are considered drug targets for aging-related diseases. The nuclear isoform Sirt1 deacetylates histones and transcription factors to regulate, e.g., metabolic adaptations and circadian mechanisms, and it is used as a therapeutic target for Huntington's disease and psoriasis. Sirt1 is regulated through a multitude of mechanisms, including the interaction with regulatory proteins such as the inhibitors Tat and Dbc1 or the activator AROS. Here, we describe a molecular characterization of AROS and how it regulates Sirt1. We find that AROS is a partly intrinsically disordered protein (IDP) that inhibits rather than activates Sirt1. A biochemical characterization of the interaction including binding and stability assays, NMR spectroscopy, mass spectrometry, and a crystal structure of Sirtuin/AROS peptide complex reveal that AROS acts as a competitive inhibitor, through binding to the Sirt1 substrate peptide site. Our results provide molecular insights in the physiological regulation of Sirt1 by a regulator protein and suggest the peptide site as an opportunity for Sirt1-targeted drug development.
- Department of Biochemistry, University of Bayreuth, 95440 Bayreuth, Germany.
Organizational Affiliation: