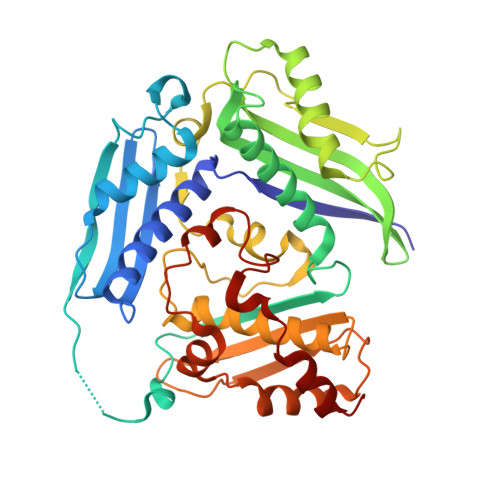
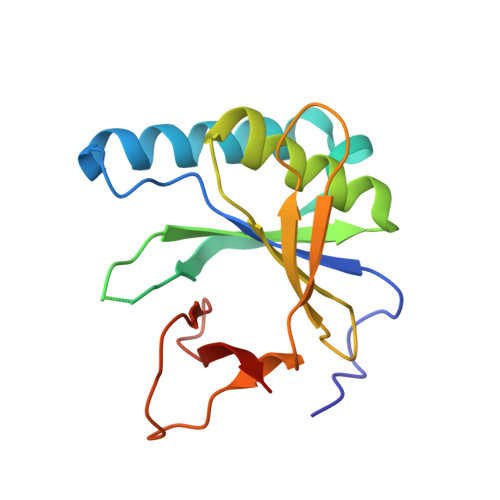
Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase.
Andriianov, A., Triguis, S., Drobiazko, A., Sierro, N., Ivanov, N.V., Selmer, M., Severinov, K., Isaev, A.(2023) Cell Rep 42: 112972-112972
- PubMed: 37578860
- DOI: https://doi.org/10.1016/j.celrep.2023.112972
- Primary Citation of Related Structures:
8BB1 - PubMed Abstract:
Bacteriophage T3 encodes a SAMase that, through cleavage of S-adenosyl methionine (SAM), circumvents the SAM-dependent type I restriction-modification (R-M) defense. We show that SAMase also allows T3 to evade the BREX defense. Although SAM depletion weakly affects BREX methylation, it completely inhibits the defensive function of BREX, suggesting that SAM could be a co-factor for BREX-mediated exclusion of phage DNA, similar to its anti-defense role in type I R-M. The anti-BREX activity of T3 SAMase is mediated not just by enzymatic degradation of SAM but also by direct inhibition of MetK, the host SAM synthase. We present a 2.8 Å cryoelectron microscopy (cryo-EM) structure of the eight-subunit T3 SAMase-MetK complex. Structure-guided mutagenesis reveals that this interaction stabilizes T3 SAMase in vivo, further stimulating its anti-BREX activity. This work provides insights in the versatility of bacteriophage counterdefense mechanisms and highlights the role of SAM as a co-factor of diverse bacterial immunity systems.
- Skolkovo Institute of Science and Technology, Moscow 143028, Russia.
Organizational Affiliation: