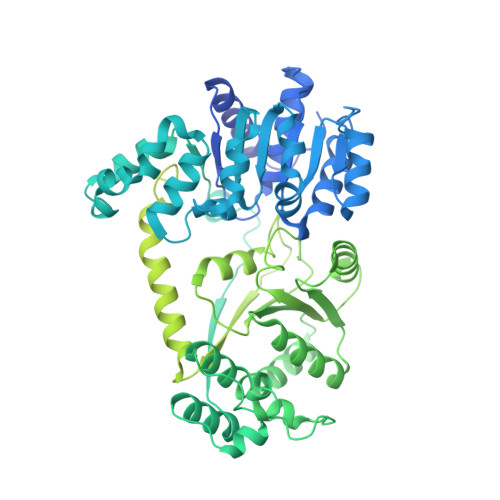
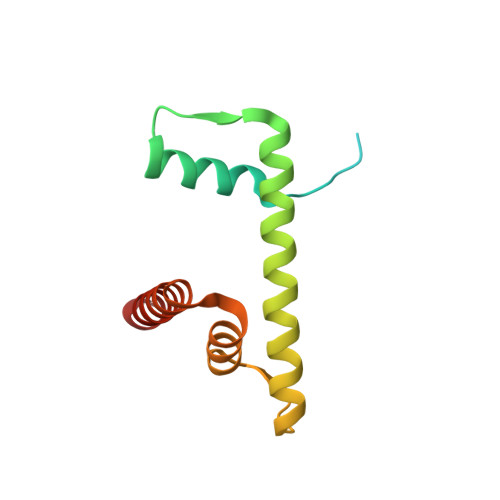
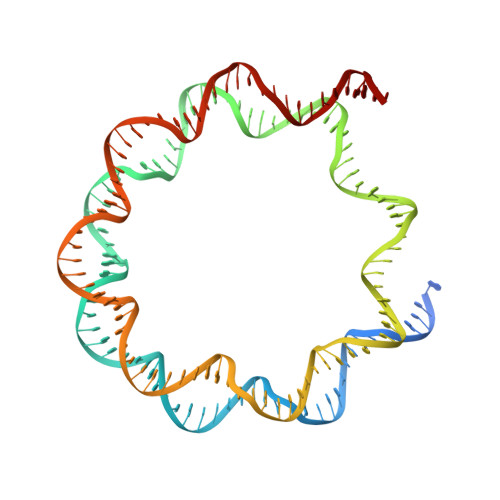
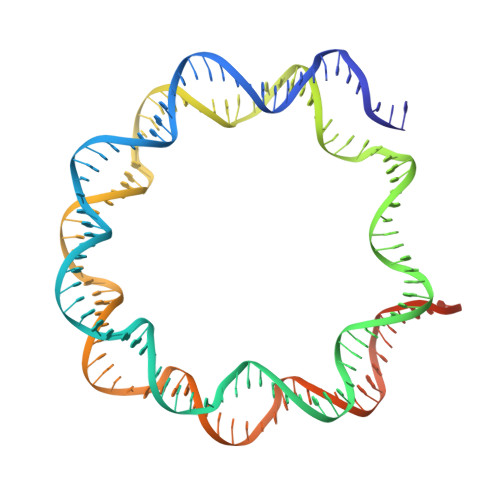
Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1.
Bacic, L., Gaullier, G., Mohapatra, J., Mao, G., Brackmann, K., Panfilov, M., Liszczak, G., Sabantsev, A., Deindl, S.(2024) Nat Commun 15: 1000-1000
- PubMed: 38307862
- DOI: https://doi.org/10.1038/s41467-024-45237-8
- Primary Citation of Related Structures:
8B0A - PubMed Abstract:
The chromatin remodeler ALC1 is activated by DNA damage-induced poly(ADP-ribose) deposited by PARP1/PARP2 and their co-factor HPF1. ALC1 has emerged as a cancer drug target, but how it is recruited to ADP-ribosylated nucleosomes to affect their positioning near DNA breaks is unknown. Here we find that PARP1/HPF1 preferentially initiates ADP-ribosylation on the histone H2B tail closest to the DNA break. To dissect the consequences of such asymmetry, we generate nucleosomes with a defined ADP-ribosylated H2B tail on one side only. The cryo-electron microscopy structure of ALC1 bound to such an asymmetric nucleosome indicates preferential engagement on one side. Using single-molecule FRET, we demonstrate that this asymmetric recruitment gives rise to directed sliding away from the DNA linker closest to the ADP-ribosylation site. Our data suggest a mechanism by which ALC1 slides nucleosomes away from a DNA break to render it more accessible to repair factors.
- Department of Cell and Molecular Biology, Science for Life Laboratory, Uppsala University, 75124, Uppsala, Sweden.
Organizational Affiliation: