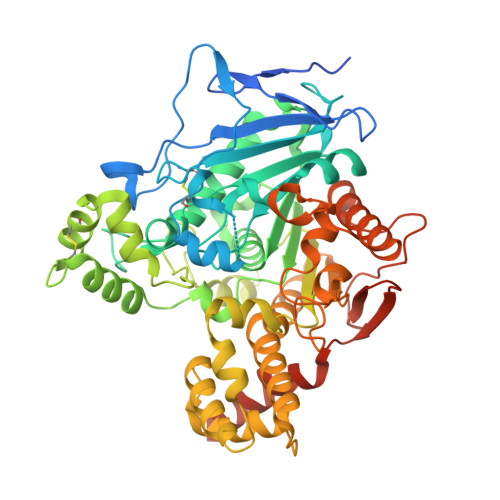
The Crystal Structure of Mouse Ces2c, a Potential Ortholog of Human CES2, Shows Structural Similarities in Substrate Regulation and Product Release to Human CES1.
Eisner, H., Riegler-Berket, L., Gamez, C.F.R., Sagmeister, T., Chalhoub, G., Darnhofer, B., Jazleena, P.J., Birner-Gruenberger, R., Pavkov-Keller, T., Haemmerle, G., Schoiswohl, G., Oberer, M.(2022) Int J Mol Sci 23
- PubMed: 36361897
- DOI: https://doi.org/10.3390/ijms232113101
- Primary Citation of Related Structures:
8AXC - PubMed Abstract:
Members of the carboxylesterase 2 (Ces2/CES2) family have been studied intensively with respect to their hydrolytic function on (pro)drugs, whereas their physiological role in lipid and energy metabolism has been realized only within the last few years. Humans have one CES2 gene which is highly expressed in liver, intestine, and kidney. Interestingly, eight homologous Ces2 (Ces2a to Ces2h) genes exist in mice and the individual roles of the corresponding proteins are incompletely understood. Mouse Ces2c (mCes2c) is suggested as potential ortholog of human CES2. Therefore, we aimed at its structural and biophysical characterization. Here, we present the first crystal structure of mCes2c to 2.12 Å resolution. The overall structure of mCes2c resembles that of the human CES1 (hCES1). The core domain adopts an α/β hydrolase-fold with S230, E347, and H459 forming a catalytic triad. Access to the active site is restricted by the cap, the flexible lid, and the regulatory domain. The conserved gate (M417) and switch (F418) residues might have a function in product release similar as suggested for hCES1. Biophysical characterization confirms that mCes2c is a monomer in solution. Thus, this study broadens our understanding of the mammalian carboxylesterase family and assists in delineating the similarities and differences of the different family members.
- Institute of Molecular Biosciences, University of Graz, 8010 Graz, Austria.
Organizational Affiliation: