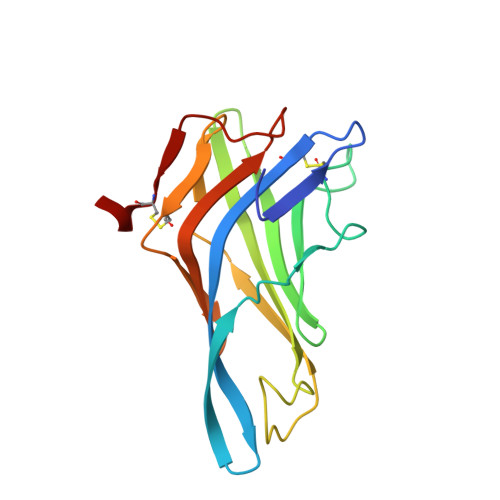
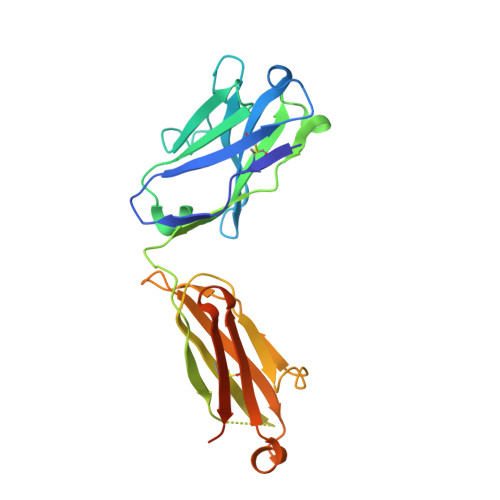
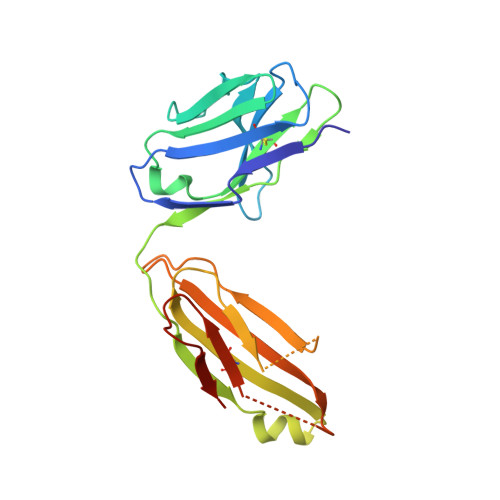
Deciphering cross-species reactivity of LAMP-1 antibodies using deep mutational epitope mapping and AlphaFold.
Pruvost, T., Mathieu, M., Dubois, S., Maillere, B., Vigne, E., Nozach, H.(2023) MAbs 15: 2175311-2175311
- PubMed: 36797224
- DOI: https://doi.org/10.1080/19420862.2023.2175311
- Primary Citation of Related Structures:
8ATH - PubMed Abstract:
Delineating the precise regions on an antigen that are targeted by antibodies has become a key step for the development of antibody therapeutics. X-ray crystallography and cryogenic electron microscopy are considered the gold standard for providing precise information about these binding sites at atomic resolution. However, they are labor-intensive and a successful outcome is not guaranteed. We used deep mutational scanning (DMS) of the human LAMP-1 antigen displayed on yeast surface and leveraged next-generation sequencing to observe the effect of individual mutants on the binding of two LAMP-1 antibodies and to determine their functional epitopes on LAMP-1. Fine-tuned epitope mapping by DMS approaches is augmented by knowledge of experimental antigen structure. As human LAMP-1 structure has not yet been solved, we used the AlphaFold predicted structure of the full-length protein to combine with DMS data and ultimately finely map antibody epitopes. The accuracy of this method was confirmed by comparing the results to the co-crystal structure of one of the two antibodies with a LAMP-1 luminal domain. Finally, we used AlphaFold models of non-human LAMP-1 to understand the lack of mAb cross-reactivity. While both epitopes in the murine form exhibit multiple mutations in comparison to human LAMP-1, only one and two mutations in the Macaca form suffice to hinder the recognition by mAb B and A, respectively. Altogether, this study promotes a new application of AlphaFold to speed up precision mapping of antibody-antigen interactions and consequently accelerate antibody engineering for optimization.
- CEA, INRAE, Medicines and Healthcare Technologies Department, Université Paris-Saclay, SIMoS, France.
Organizational Affiliation: