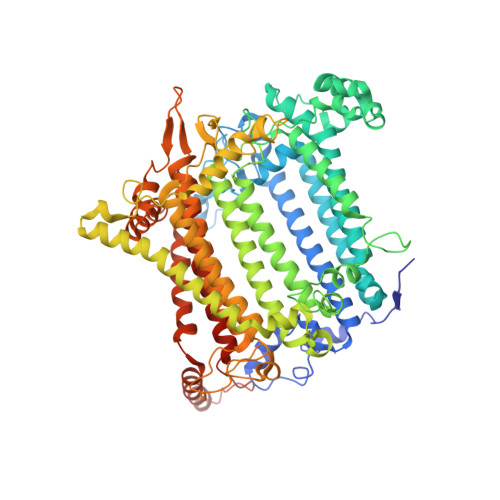
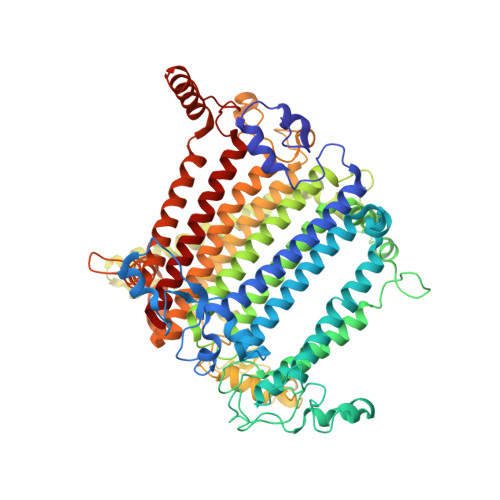
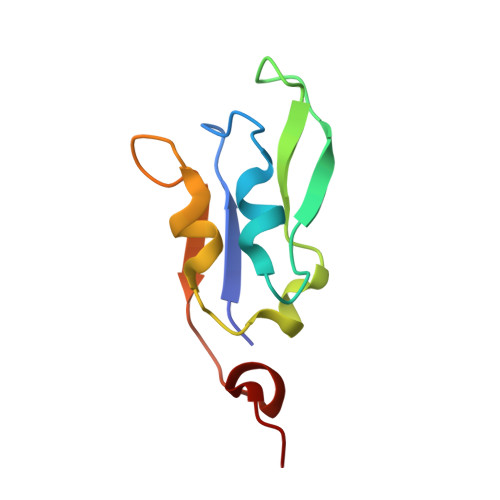
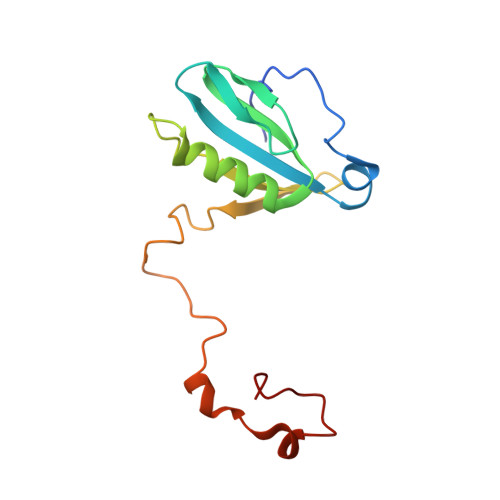
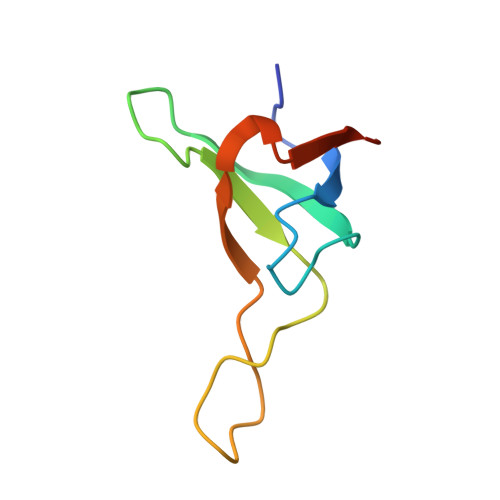
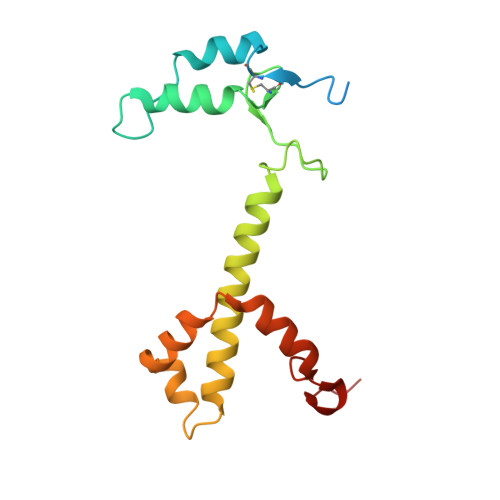
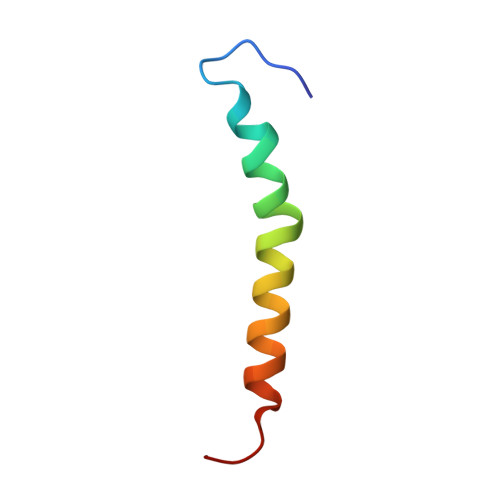
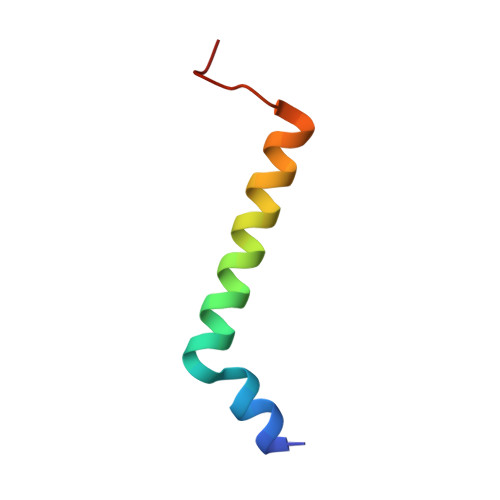
The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Zhao, Z., Vercellino, I., Knoppova, J., Sobotka, R., Murray, J.W., Nixon, P.J., Sazanov, L.A., Komenda, J.(2023) Nat Commun 14: 4681-4681
- PubMed: 37542031
- DOI: https://doi.org/10.1038/s41467-023-40388-6
- Primary Citation of Related Structures:
8AM5, 8ASL, 8ASP - PubMed Abstract:
Robust oxygenic photosynthesis requires a suite of accessory factors to ensure efficient assembly and repair of the oxygen-evolving photosystem two (PSII) complex. The highly conserved Ycf48 assembly factor binds to the newly synthesized D1 reaction center polypeptide and promotes the initial steps of PSII assembly, but its binding site is unclear. Here we use cryo-electron microscopy to determine the structure of a cyanobacterial PSII D1/D2 reaction center assembly complex with Ycf48 attached. Ycf48, a 7-bladed beta propeller, binds to the amino-acid residues of D1 that ultimately ligate the water-oxidising Mn 4 CaO 5 cluster, thereby preventing the premature binding of Mn 2+ and Ca 2+ ions and protecting the site from damage. Interactions with D2 help explain how Ycf48 promotes assembly of the D1/D2 complex. Overall, our work provides valuable insights into the early stages of PSII assembly and the structural changes that create the binding site for the Mn 4 CaO 5 cluster.
- Sir Ernst Chain Building-Wolfson Laboratories, Department of Life Sciences, Imperial College London, S. Kensington Campus, London, SW7 2AZ, UK.
Organizational Affiliation: