LSSmScarlet2 and LSSmScarlet3, Chemically Stable Genetically Encoded Red Fluorescent Proteins with a Large Stokes' Shift.
Subach, O.M., Vlaskina, A.V., Agapova, Y.K., Piatkevich, K.D., Patrushev, M.V., Samygina, V.R., Subach, F.V.(2022) Int J Mol Sci 23
- PubMed: 36232354
- DOI: https://doi.org/10.3390/ijms231911051
- Primary Citation of Related Structures:
8ARM - PubMed Abstract:
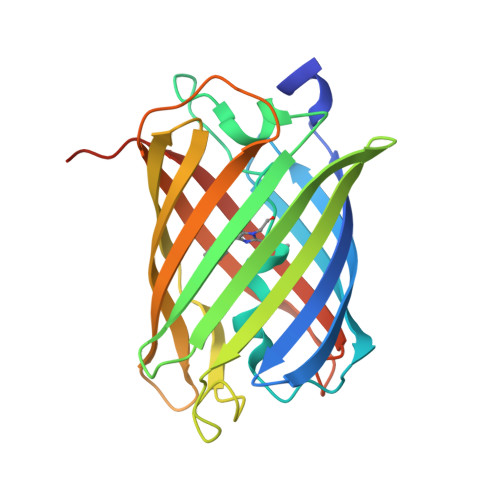
Red fluorescent proteins with a large Stokes' shift (LSSRFPs) are genetically encoded and efficiently excited by 488 nm light, allowing simultaneous dual-color one- and two-photon fluorescence imaging and fluorescence correlation spectroscopy in combination with green fluorescent proteins FPs. Recently, based on the conventional bright mScarlet RFP, we developed the LSSRFP LSSmScarlet. LSSmScarlet is characterized by two pKa values at pH values of 1.9 and 5.8. In this study, we developed improved versions of LSSmScarlet, named LSSmScarlet2 and LSSmScarlet3, which are characterized by a Stokes' shift of 128 nm and extreme pH stability with a single pKa value of 2.2. LSSmScarlet2 and LSSmScarlet3 had 1.8-fold faster and 3-fold slower maturation than LSSmScarlet, respectively. In addition, both LSSRFPs were 1.5- to 1.6-fold more photostable and more chemically resistant to denaturation by guanidinium chloride and guanidinium thiocyanate. We also compared the susceptibility of the LSSmScarlet2, LSSmScarlet3, and other LSSRFPs to the reagents used for whole-mount imaging, expansion microscopy, and immunostaining techniques. Due to higher pH stability and faster maturation, the LSSmScarlet3-LAMP3 fusion was 2.2-fold brighter than LSSmScarlet-LAMP3 in lysosomes of mammalian cells. The LSSmScarlet3-hLAMP2A fusion was similar in brightness to LSSmScarlet-hLAMP2A in lysosomes. We successfully applied the monomeric LSSmScarlet2 and LSSmScarlet3 proteins for confocal imaging of structural proteins in live mammalian cells. We also solved the X-ray structure of the LSSmScarlet2 protein at a resolution of 1.41 Å. Site-directed mutagenesis of the LSSmScarlet2 protein demonstrated the key role of the T74 residue in improving the pH and chemical stability of the LSSmScarlet2 protein.
- Complex of NBICS Technologies, National Research Center "Kurchatov Institute", 123182 Moscow, Russia.
Organizational Affiliation: