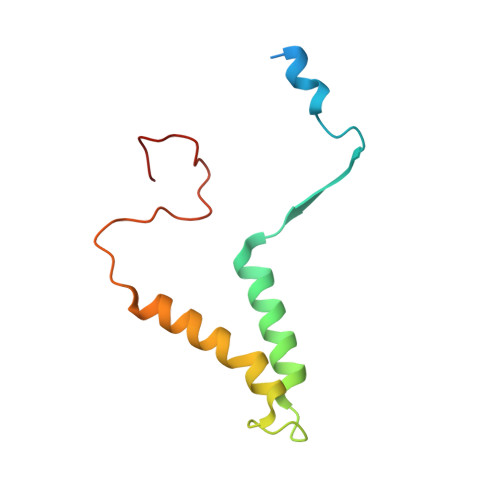
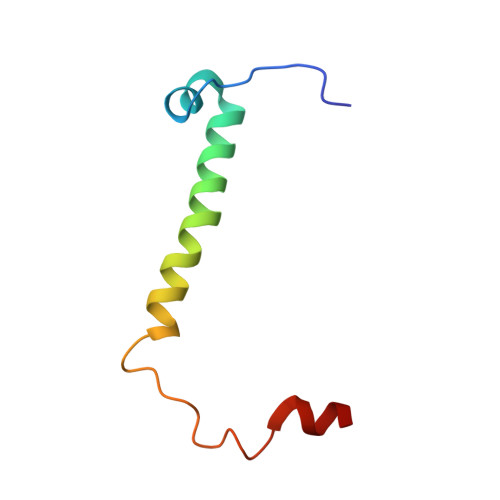
An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Gahura, O., Muhleip, A., Hierro-Yap, C., Panicucci, B., Jain, M., Hollaus, D., Slapnickova, M., Zikova, A., Amunts, A.(2022) Nat Commun 13: 5989-5989
- PubMed: 36220811
- DOI: https://doi.org/10.1038/s41467-022-33588-z
- Primary Citation of Related Structures:
8AP6, 8AP7, 8AP8, 8AP9, 8APA, 8APB, 8APC, 8APD, 8APE, 8APF, 8APG, 8APH, 8APJ, 8APK - PubMed Abstract:
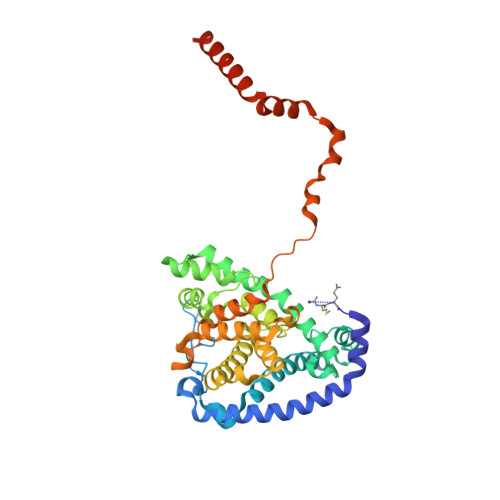
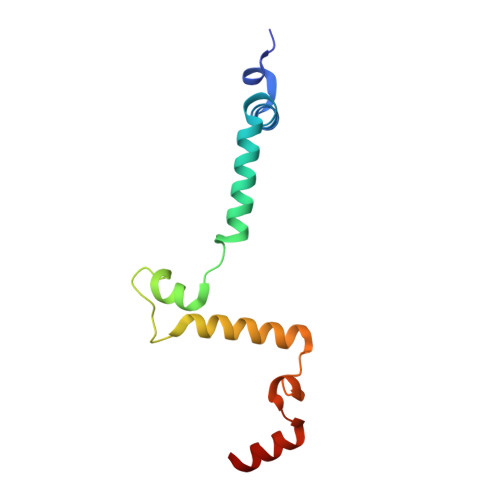
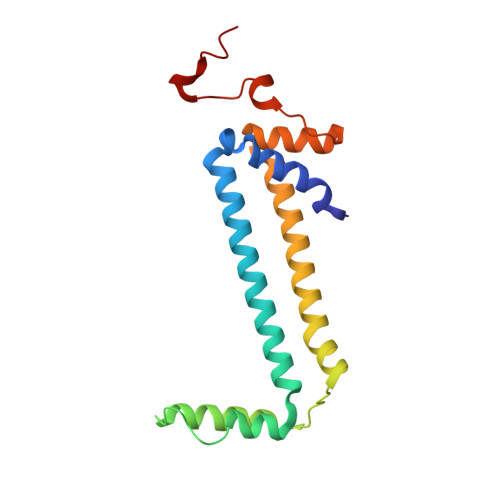
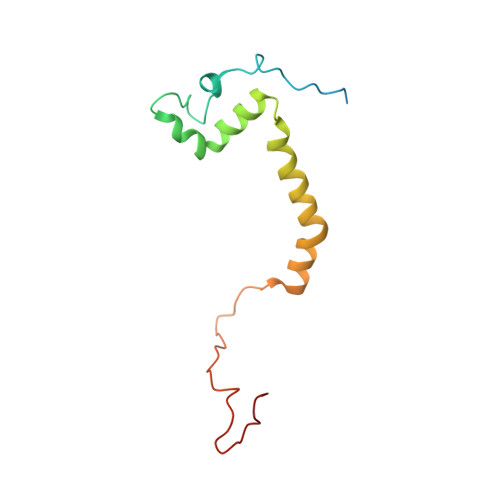
Mitochondrial ATP synthase forms stable dimers arranged into oligomeric assemblies that generate the inner-membrane curvature essential for efficient energy conversion. Here, we report cryo-EM structures of the intact ATP synthase dimer from Trypanosoma brucei in ten different rotational states. The model consists of 25 subunits, including nine lineage-specific, as well as 36 lipids. The rotary mechanism is influenced by the divergent peripheral stalk, conferring a greater conformational flexibility. Proton transfer in the lumenal half-channel occurs via a chain of five ordered water molecules. The dimerization interface is formed by subunit-g that is critical for interactions but not for the catalytic activity. Although overall dimer architecture varies among eukaryotes, we find that subunit-g together with subunit-e form an ancestral oligomerization motif, which is shared between the trypanosomal and mammalian lineages. Therefore, our data defines the subunit-g/e module as a structural component determining ATP synthase oligomeric assemblies.
- Institute of Parasitology, Biology Centre, Czech Academy of Sciences, 37005, České Budějovice, Czech Republic.
Organizational Affiliation: