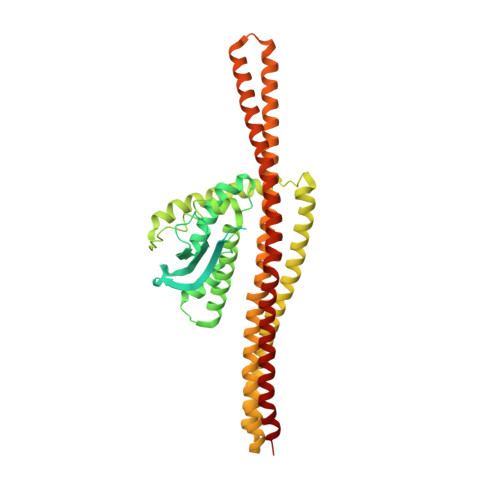
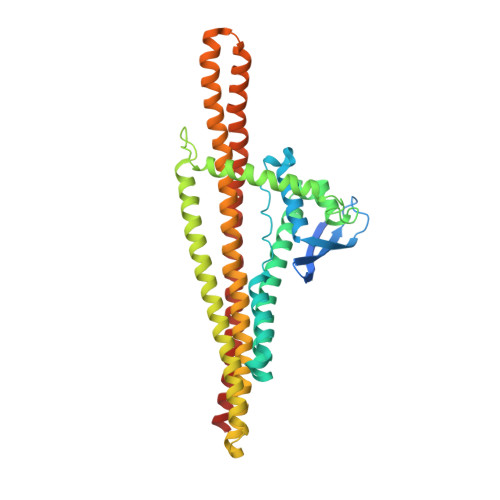
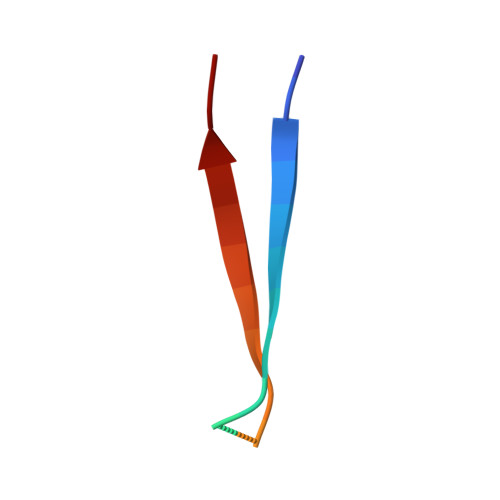
Architecture of the ESCPE-1 membrane coat.
Lopez-Robles, C., Scaramuzza, S., Astorga-Simon, E.N., Ishida, M., Williamson, C.D., Banos-Mateos, S., Gil-Carton, D., Romero-Durana, M., Vidaurrazaga, A., Fernandez-Recio, J., Rojas, A.L., Bonifacino, J.S., Castano-Diez, D., Hierro, A.(2023) Nat Struct Mol Biol 30: 958-969
- PubMed: 37322239
- DOI: https://doi.org/10.1038/s41594-023-01014-7
- Primary Citation of Related Structures:
8A1G, 8ABQ, 8AFZ - PubMed Abstract:
Recycling of membrane proteins enables the reuse of receptors, ion channels and transporters. A key component of the recycling machinery is the endosomal sorting complex for promoting exit 1 (ESCPE-1), which rescues transmembrane proteins from the endolysosomal pathway for transport to the trans-Golgi network and the plasma membrane. This rescue entails the formation of recycling tubules through ESCPE-1 recruitment, cargo capture, coat assembly and membrane sculpting by mechanisms that remain largely unknown. Herein, we show that ESCPE-1 has a single-layer coat organization and suggest how synergistic interactions between ESCPE-1 protomers, phosphoinositides and cargo molecules result in a global arrangement of amphipathic helices to drive tubule formation. Our results thus define a key process of tubule-based endosomal sorting.
- CIC bioGUNE, Derio, Spain.
Organizational Affiliation: