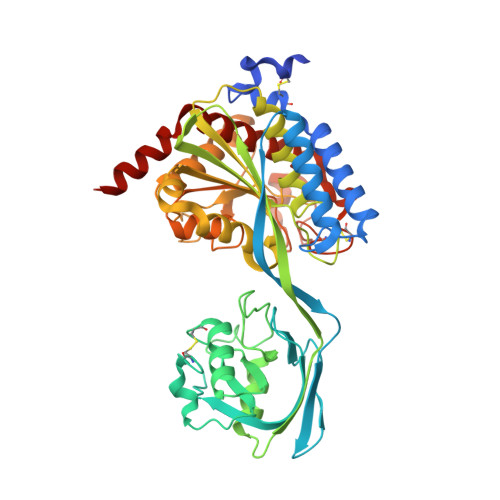
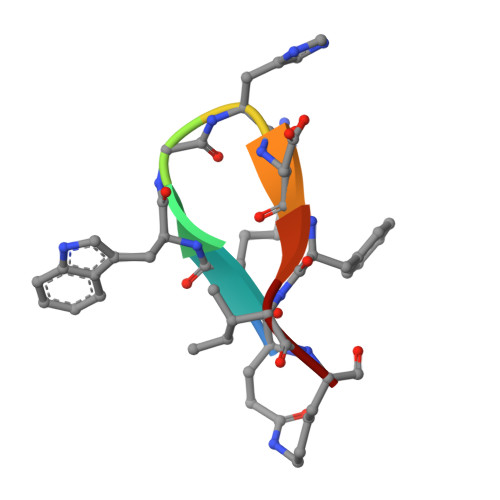
An anti-biofilm cyclic peptide targets a secreted aminopeptidase from P. aeruginosa.
Harding, C.J., Bischoff, M., Bergkessel, M., Czekster, C.M.(2023) Nat Chem Biol 19: 1158-1166
- PubMed: 37386135
- DOI: https://doi.org/10.1038/s41589-023-01373-8
- Primary Citation of Related Structures:
8AC7, 8AC9, 8ACG, 8ACK, 8ACR - PubMed Abstract:
Pseudomonas aeruginosa is an opportunistic pathogen that causes serious illness, especially in immunocompromised individuals. P. aeruginosa forms biofilms that contribute to growth and persistence in a wide range of environments. Here we investigated the aminopeptidase, P. aeruginosa aminopeptidase (PaAP) from P. aeruginosa, which is highly abundant in the biofilm matrix. PaAP is associated with biofilm development and contributes to nutrient recycling. We confirmed that post-translational processing was required for activation and PaAP is a promiscuous aminopeptidase acting on unstructured regions of peptides and proteins. Crystal structures of wild-type enzymes and variants revealed the mechanism of autoinhibition, whereby the C-terminal propeptide locks the protease-associated domain and the catalytic peptidase domain into a self-inhibited conformation. Inspired by this, we designed a highly potent small cyclic-peptide inhibitor that recapitulates the deleterious phenotype observed with a PaAP deletion variant in biofilm assays and present a path toward targeting secreted proteins in a biofilm context.
- Biomedical Sciences Research Complex, School of Biology, University of St Andrews, St Andrews, UK.
Organizational Affiliation: