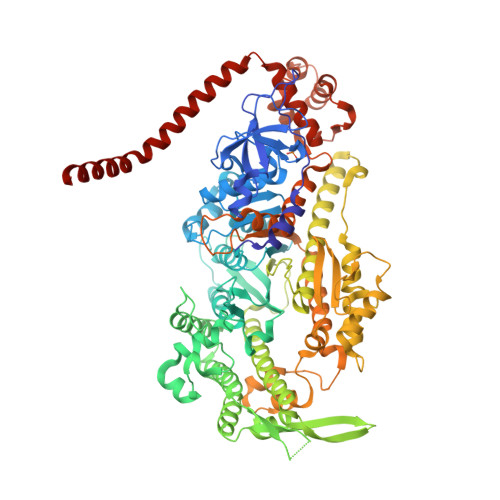
Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Moussaoui, D., Robblee, J.P., Robert-Paganin, J., Auguin, D., Fisher, F., Fagnant, P.M., Macfarlane, J.E., Schaletzky, J., Wehri, E., Mueller-Dieckmann, C., Baum, J., Trybus, K.M., Houdusse, A.(2023) Nat Commun 14: 3463-3463
- PubMed: 37308472
- DOI: https://doi.org/10.1038/s41467-023-38976-7
- Primary Citation of Related Structures:
8A12, 8CDM, 8CDQ - PubMed Abstract:
Malaria results in more than 500,000 deaths per year and the causative Plasmodium parasites continue to develop resistance to all known agents, including different antimalarial combinations. The class XIV myosin motor PfMyoA is part of a core macromolecular complex called the glideosome, essential for Plasmodium parasite mobility and therefore an attractive drug target. Here, we characterize the interaction of a small molecule (KNX-002) with PfMyoA. KNX-002 inhibits PfMyoA ATPase activity in vitro and blocks asexual blood stage growth of merozoites, one of three motile Plasmodium life-cycle stages. Combining biochemical assays and X-ray crystallography, we demonstrate that KNX-002 inhibits PfMyoA using a previously undescribed binding mode, sequestering it in a post-rigor state detached from actin. KNX-002 binding prevents efficient ATP hydrolysis and priming of the lever arm, thus inhibiting motor activity. This small-molecule inhibitor of PfMyoA paves the way for the development of alternative antimalarial treatments.
- Structural Motility, Institut Curie, Université Paris Sciences et Lettres, Sorbonne Université, CNRS UMR144, 75248, Paris, France.
Organizational Affiliation: