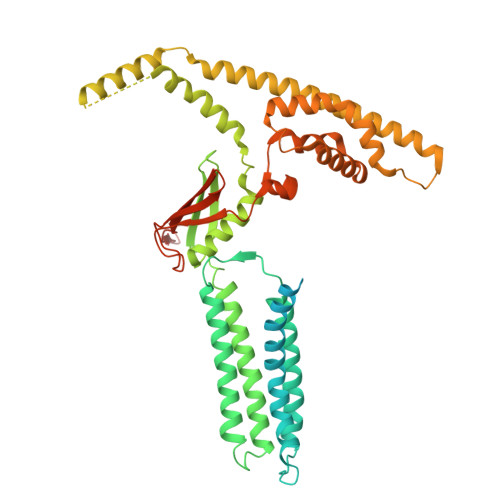
Cryo-EM structure and oligomerization of the human planar cell polarity core protein Vangl1.
Zhang, F., Li, S., Wu, H., Chen, S.(2025) Nat Commun 16: 135-135
- PubMed: 39753546
- DOI: https://doi.org/10.1038/s41467-024-55397-2
- Primary Citation of Related Structures:
8ZXD - PubMed Abstract:
Vangl is a planar cell polarity (PCP) core protein essential for aligned cell orientation along the epithelial plane perpendicular to the apical-basal direction, which is important for tissue morphogenesis, development and collective cell behavior. Mutations in Vangl are associated with developmental defects, including neural tube defects (NTDs), according to human cohort studies of sporadic and familial cases. The complex mechanisms underlying Vangl-mediated PCP signaling or Vangl-associated human congenital diseases have been hampered by the lack of molecular characterizations of Vangl. Here, we show biochemical and structural evidence that human Vangl1 oligomerizes as dimers of trimers, and that the dimerization of trimers promotes binding to the PCP effector Prickle1 (Pk1) in vitro. Mapping of human disease-associated point mutations suggests potential pathological mechanisms and paves the way for future studies on the importance of lipid binding, central vestibule and oligomerization of Vangl, thereby providing insights into the molecular mechanisms of the PCP signaling pathway.
- Shanghai Institute of Precision Medicine, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Organizational Affiliation: