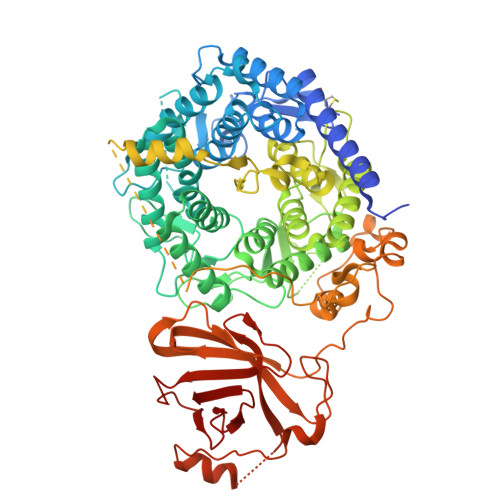
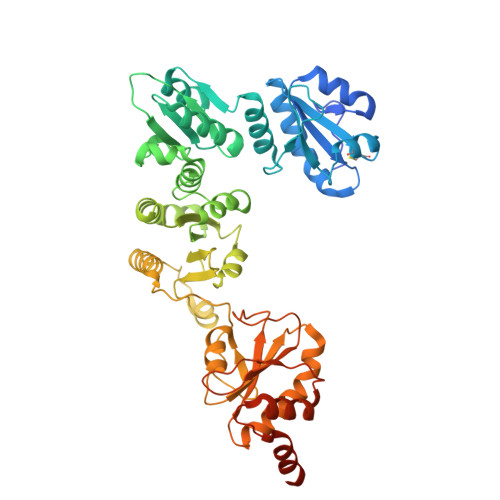
Initiation of ERAD by the bifunctional complex of Mnl1/Htm1 mannosidase and protein disulfide isomerase.
Zhao, D., Wu, X., Rapoport, T.A.(2025) Nat Struct Mol Biol 32: 1006-1018
- PubMed: 39930008
- DOI: https://doi.org/10.1038/s41594-025-01491-y
- Primary Citation of Related Structures:
8ZPW - PubMed Abstract:
Misfolded glycoproteins in the endoplasmic reticulum (ER) lumen are translocated into the cytosol and degraded by the proteasome, a conserved process called ER-associated protein degradation (ERAD). In Saccharomyces cerevisiae, the glycan of these proteins is trimmed by the luminal mannosidase Mnl1 (Htm1) to generate a degradation signal. Interestingly, Mnl1 is associated with protein disulfide isomerase (Pdi1). Here we used cryo-electron microscopy, biochemical and in vivo experiments to elucidate how this complex initiates ERAD. The Mnl1-Pdi1 complex first demannosylates misfolded, globular proteins that are recognized through the C-terminal domain (CTD) of Mnl1; Pdi1 causes the CTD to ignore completely unfolded polypeptides. The disulfides of these globular proteins are then reduced by the Pdi1 component of the complex. Mnl1 blocks the canonical oxidative function of Pdi1, allowing it to function as a disulfide reductase in ERAD. The generated unfolded polypeptides can then be translocated across the membrane into the cytosol.
- Howard Hughes Medical Institute and Department of Cell Biology, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: