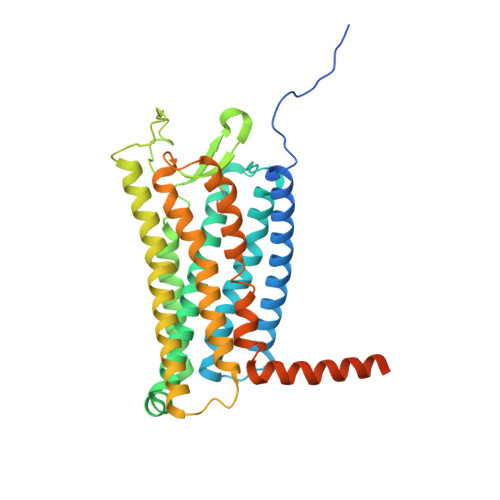
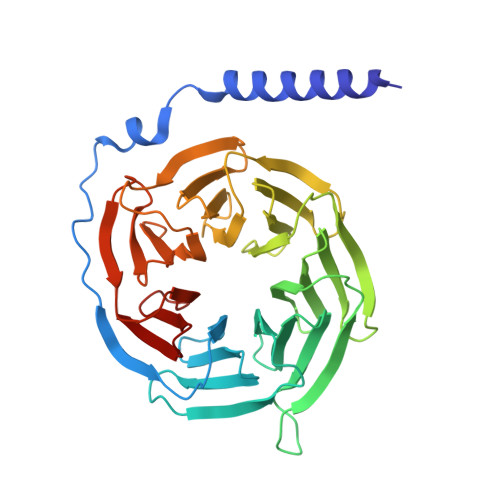
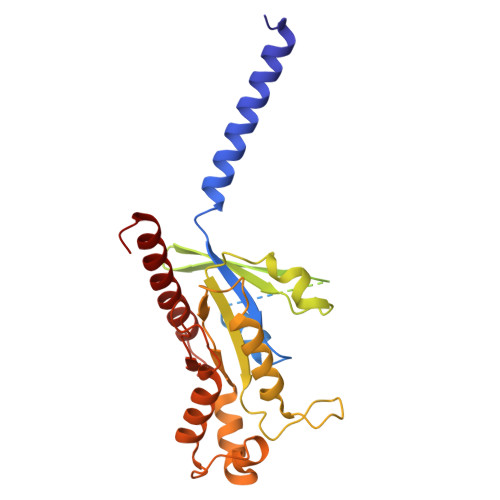
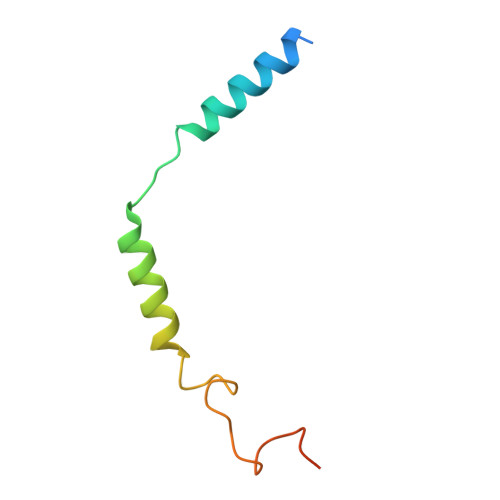
Structural basis for full-length chemerin recognition and signaling through chemerin receptor 1.
Liu, A., Liu, Y., Wang, J., Ye, R.D.(2024) Commun Biol 7: 1598-1598
- PubMed: 39616240
- DOI: https://doi.org/10.1038/s42003-024-07228-9
- Primary Citation of Related Structures:
8ZJG - PubMed Abstract:
Chemerin, a chemotactic adipokine, plays essential roles in adipogenesis and inflammation. Serum chemerin concentration is closely associated with obesity and metabolism disorders. The mature form of chemerin (residues 21-157) acts primarily through chemerin receptor 1 (CMKLR1) for transmembrane signaling. As a result, CMKLR1 serves as a promising target for therapeutic intervention of immunometabolic diseases such as diabetes and multiple sclerosis. Here, we present a high-resolution cryo-EM structure of CMKLR1-Gi signaling complex bound to biologically active full-length chemerin. The mature chemerin shows binding features distinct from its C-terminal nonapeptide including interaction with both the extracellular loops (ECLs) and the N-terminus of CMKLR1. Combining results from functional assays, our studies demonstrate that chemerin interacts with CMKLR1 in a "two-site" mode similar to chemokine-chemokine receptor interactions, but acting as a "reverse chemokine" by inserting its C-terminus instead of the N-terminus as in the case of chemokines into the transmembrane binding pocket of CMKLR1. These structural insights are expected to help develop synthetic analogs with therapeutic potential.
- Dongguan Songshan Lake Central Hospital, Dongguan Third People's Hospital, The Affiliated Dongguan Songshan Lake Central Hospital, Guangdong Medical University, Dongguan, Guangdong, 523326, China. liuaijun@cuhk.edu.cn.
Organizational Affiliation: