Structure of trypsinogen-engaged EP at 2.95 angstroms resolution
Song, Q.Y., Ding, Z.Y., Huang, H.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine protease 1 | A [auth C] | 232 | Homo sapiens | Mutation(s): 0 EC: 3.4.21.4 | 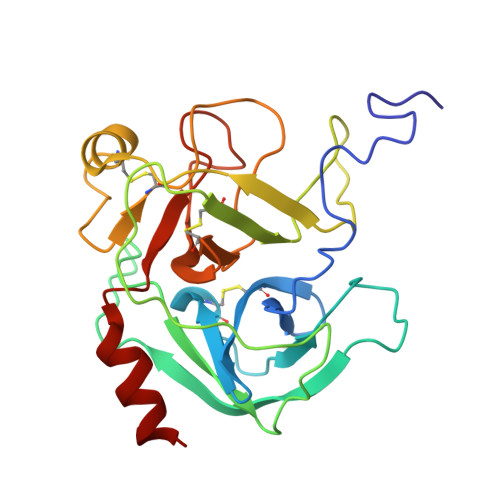 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P07477 (Homo sapiens) Explore P07477 Go to UniProtKB: P07477 | |||||
PHAROS: P07477 GTEx: ENSG00000204983 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07477 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Enteropeptidase catalytic light chain | B [auth D] | 235 | Homo sapiens | Mutation(s): 3 Gene Names: TMPRSS15, ENTK, PRSS7 EC: 3.4.21.9 | 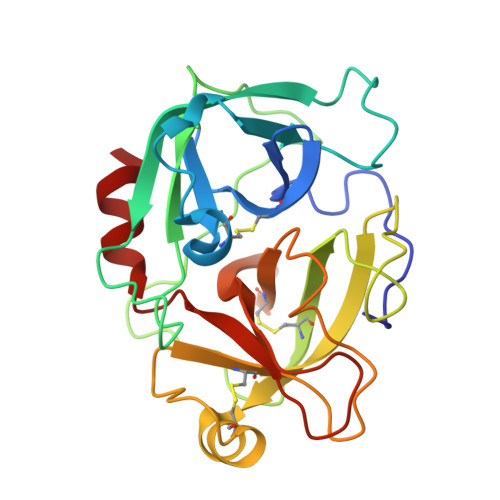 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P98073 (Homo sapiens) Explore P98073 Go to UniProtKB: P98073 | |||||
PHAROS: P98073 GTEx: ENSG00000154646 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P98073 | ||||
Glycosylation | |||||
| Glycosylation Sites: 5 | Go to GlyGen: P98073-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Enteropeptidase non-catalytic heavy chain | C [auth A] | 603 | Homo sapiens | Mutation(s): 0 Gene Names: TMPRSS15, ENTK, PRSS7 EC: 3.4.21.9 | 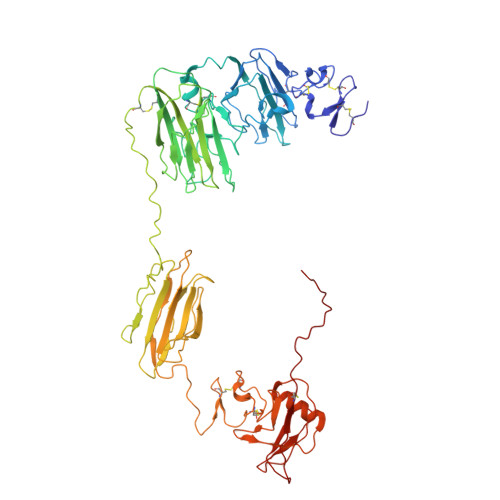 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P98073 (Homo sapiens) Explore P98073 Go to UniProtKB: P98073 | |||||
PHAROS: P98073 GTEx: ENSG00000154646 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P98073 | ||||
Glycosylation | |||||
| Glycosylation Sites: 11 | Go to GlyGen: P98073-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | D [auth B], E, L, M, N, D [auth B], E, L, M, N, O | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G15407YE GlyCosmos: G15407YE GlyGen: G15407YE | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | F, G | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | H, K | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22573RC GlyCosmos: G22573RC GlyGen: G22573RC | |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | I | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G73474XW GlyCosmos: G73474XW GlyGen: G73474XW | |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | J | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G81315DD GlyCosmos: G81315DD GlyGen: G81315DD | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | P [auth D], Q [auth A], R [auth A], S [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |