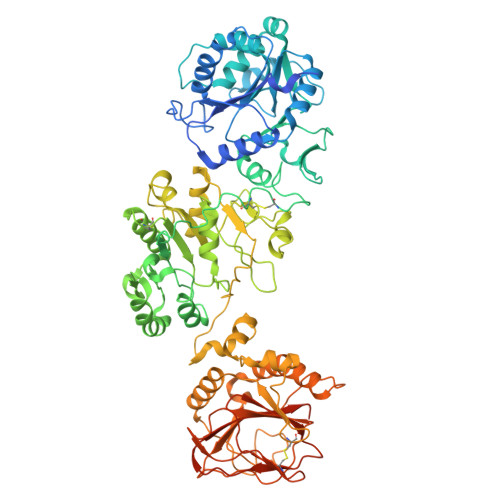
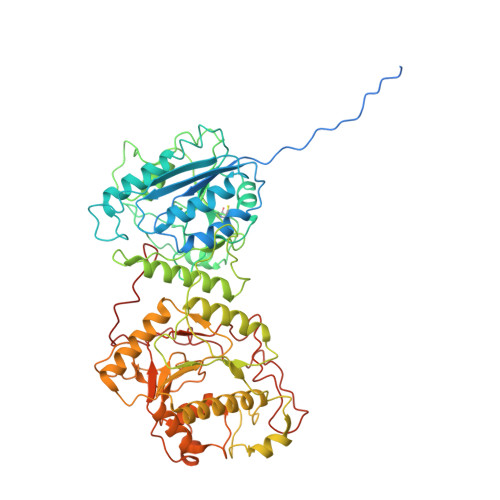
The structural basis for the human procollagen lysine hydroxylation and dual-glycosylation.
Peng, J., Li, W., Yao, D., Xia, Y., Wang, Q., Cai, Y., Li, S., Cao, M., Shen, Y., Ma, P., Liao, R., Zhao, J., Qin, A., Cao, Y.(2025) Nat Commun 16: 2436-2436
- PubMed: 40069201
- DOI: https://doi.org/10.1038/s41467-025-57768-9
- Primary Citation of Related Structures:
8ZGC, 8ZGE, 8ZGG, 8ZGH - PubMed Abstract:
The proper assembly and maturation of collagens necessitate the orchestrated hydroxylation and glycosylation of multiple lysyl residues in procollagen chains. Dysfunctions in this multistep modification process can lead to severe collagen-associated diseases. To elucidate the coordination of lysyl processing activities, we determine the cryo-EM structures of the enzyme complex formed by LH3/PLOD3 and GLT25D1/ColGalT1, designated as the KOGG complex. Our structural analysis reveals a tetrameric complex comprising dimeric LH3/PLOD3s and GLT25D1/ColGalT1s, assembled with interactions involving the N-terminal loop of GLT25D1/ColGalT1 bridging another GLT25D1/ColGalT1 and LH3/PLOD3. We further elucidate the spatial configuration of the hydroxylase, galactosyltransferase, and glucosyltransferase sites within the KOGG complex, along with the key residues involved in substrate binding at these enzymatic sites. Intriguingly, we identify a high-order oligomeric pattern characterized by the formation of a fiber-like KOGG polymer assembled through the repetitive incorporation of KOGG tetramers as the biological unit.
- Department of Orthopaedics, Shanghai Key Laboratory of Orthopaedic Implant, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200011, China.
Organizational Affiliation: