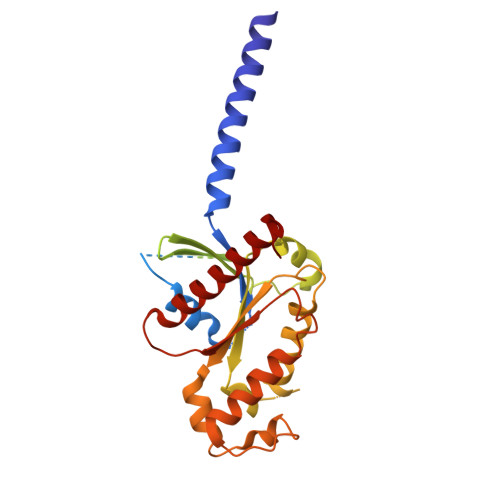
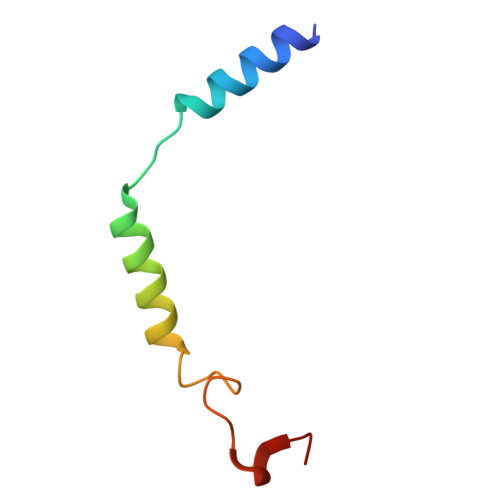
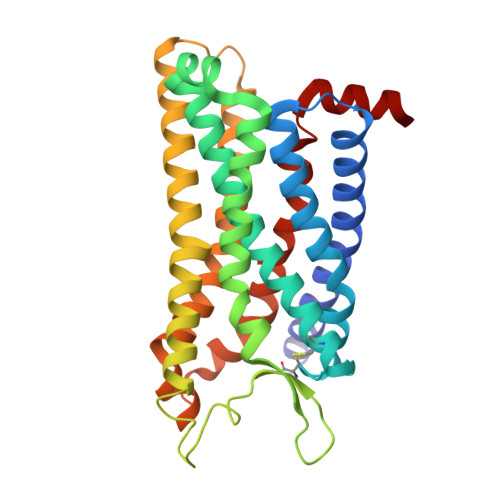
Oral FPR2/ALX modulators tune myeloid cell activity to ameliorate mucosal inflammation in inflammatory bowel disease.
Yang, W.S., Liu, Q., Li, Y., Li, G.Y., Lin, S., Li, J., Li, L.Y., Li, Y., Ge, X.L., Wang, X.Z., Wu, W., Yan, J., Wang, G.F., Zhou, Q.T., Liu, Q., Wang, M.W., Li, Z.P.(2025) Acta Pharmacol Sin 46: 1958-1973
- PubMed: 40069490
- DOI: https://doi.org/10.1038/s41401-025-01525-7
- Primary Citation of Related Structures:
8ZBW - PubMed Abstract:
Current treatments of inflammatory bowel disease (IBD) largely depend on anti-inflammatory and immunosuppressive strategies with unacceptable efficacy and adverse events. Resolution or repair agents to treat IBD are not available but potential targets like formyl peptide receptor 2 (FPR2/ALX) may fill the gap. In this study we evaluated the therapeutic effects of two small molecule FPR2/ALX modulators (agonist Quin-C1 and antagonist Quin-C7) against IBD. We first analyzed the cryo-electron microscopy structure of the Quin-C1-FPR2 in complex with heterotrimeric G i to reveal the structural basis for ligand recognition and FPR2 activation. We then established dextran sulfate sodium (DSS)-induced colitis model in both normal and myeloid depletion mice. We showed that oral administration of Quin-C1 for 7 days ameliorated DSS-induced colitis evidenced by alleviated disease activity indexes, reduced colonic histopathological scores, and corrected cytokine disorders. Meanwhile, we found that oral administration of FPR2/ALX antagonist Quin-C7 exerted therapeutic actions similar to those of Quin-C1. In terms of symptomatic improvements, the ED 50 values of Quin-C1 and Quin-C7 were 1.3660 mg/kg and 2.2110 mg/kg, respectively. The underlying mechanisms involved ERK- or ERK/JNK-mediated myeloid cell regulation that limited the development of colitis and inflammation. This is the first demonstration of anti-colitis property caused by synthetic small molecule FPR2/ALX modulators, implying that FPR2/ALX modulation rather than agonism alone ameliorates IBD.
- Department of Clinical Pharmacy, Children's Hospital of Fudan University, National Children's Medical Center, Shanghai, 201102, China.
Organizational Affiliation: