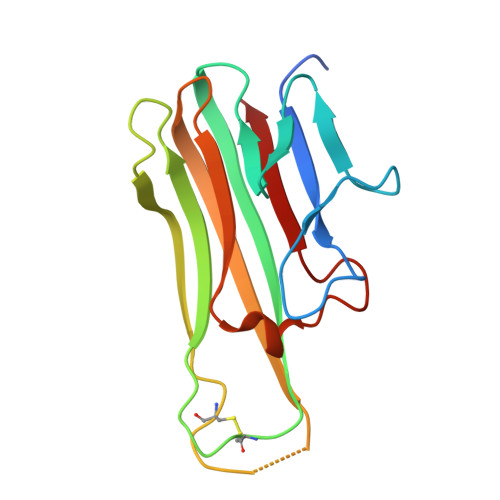
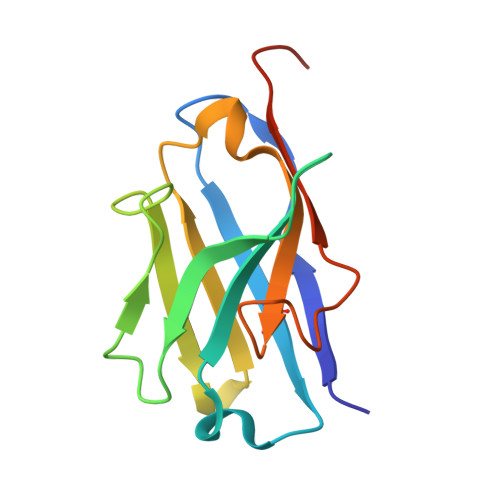
Structural design of the anti-TNF alpha therapeutic NANOBODY® compound, ozoralizumab, to support its potent and sustained clinical efficacy.
Mima, M., Mishima-Tsumagari, C., Nakano, K., Morimoto, M., Ogata, H., Sakata, M., Iwaoka, R., Iwata, K., Hachiuma, K., Iwamoto, K., Fujii, Y., Kurokawa, T.(2024) Biochem Biophys Res Commun 734: 150454-150454
- PubMed: 39083975
- DOI: https://doi.org/10.1016/j.bbrc.2024.150454
- Primary Citation of Related Structures:
8Z8M, 8Z8V - Taisho Pharmaceutical Co., Ltd., 1-403, Yoshino-cho, Kita-ku, Saitama, 331-9530, Japan.
Organizational Affiliation: