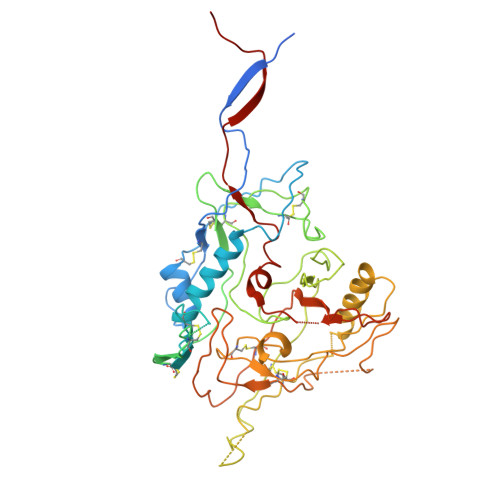
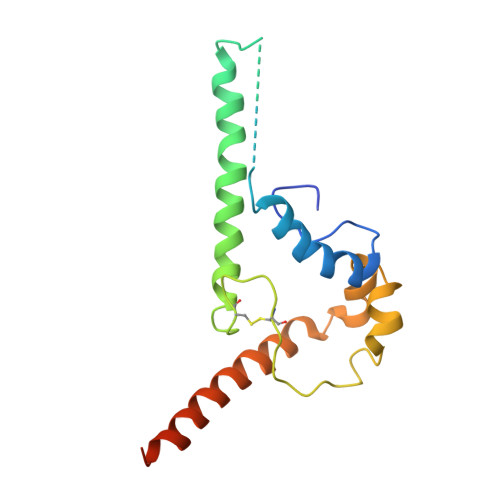
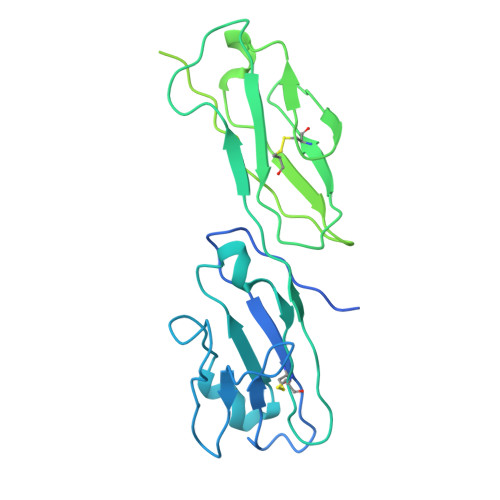
Intermediate open state of CD4-bound HIV-1 env heterotrimers in asia CRFs.
Li, D., Liu, L., Ye, X., Chen, Y., Ren, Q., Xu, S., Ren, Y., Cao, H., Wang, T.(2024) Biochem Biophys Res Commun 725: 150249-150249
- PubMed: 38880081
- DOI: https://doi.org/10.1016/j.bbrc.2024.150249
- Primary Citation of Related Structures:
8Z7N - PubMed Abstract:
The HIV-1 envelope glycoprotein (Env) plays crucial role in viral infection by facilitating viral attachment to host cells and inducing fusion of the virus with the host cell membrane. This fusion allows the HIV-1 viral genome to enter the target cell then triggering various stages of the viral life cycle. The native Env directly interacts with the main receptor CD4 and the co-receptor (CCR5 or CXCR4) in human cell membrane then induces membrane fusion. The elucidation of the structure of Env with CD4 and co-receptors in different HIV-1 subtypes is essential for the understanding of the mechanism of virus entry. Here we report the Cryo-EM structure of the CD4-bound HIV-1 heterotrimeric Env from Asia prevalent CRF07_BC CH119 strain. In this structure, the binding of three CD4 molecules with Env induced extensively conformational changes in gp120, resulting in the transformation of the Env from close state to intermediate open state. Additionally, the conformational shift of V1/V2 loops of the heterotrimeric Env allosterically expose the V3 loop and promoting the further interactions with co-receptor CCR5 or CXCR4. These findings not only illustrate the structural complexity and plasticity of HIV-1 Env but also give new insights how the biological trimeric Env initialize the immune recognition and membrane fusion.
- School of basic medical Sciences, Capital Medical University, 10 Xitoutiao You'anMen Street, Beijing, 100069, China; Institute of Infectious Diseases, Shenzhen Bay Laboratory, Guangming District, Shenzhen, 518132, China.
Organizational Affiliation: