Structure of polycystin-1/polycystin-2 complex with PI(4)P-bound
Chen, M., Su, Q., Shi, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polycystin-1 | 1,261 | Homo sapiens | Mutation(s): 0 Gene Names: PKD1 | 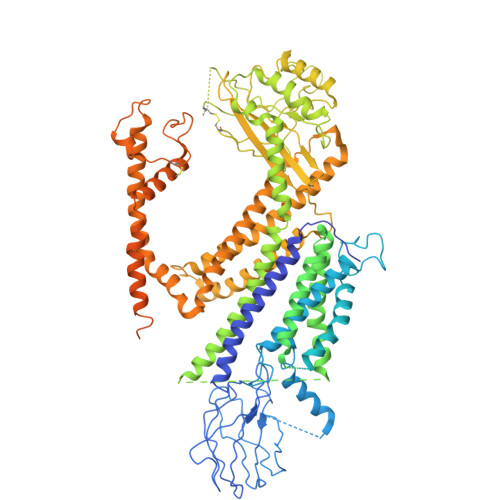 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P98161 (Homo sapiens) Explore P98161 Go to UniProtKB: P98161 | |||||
PHAROS: P98161 GTEx: ENSG00000008710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P98161 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polycystin-2 | 1,007 | Homo sapiens | Mutation(s): 0 Gene Names: PKD2, TRPP2 | 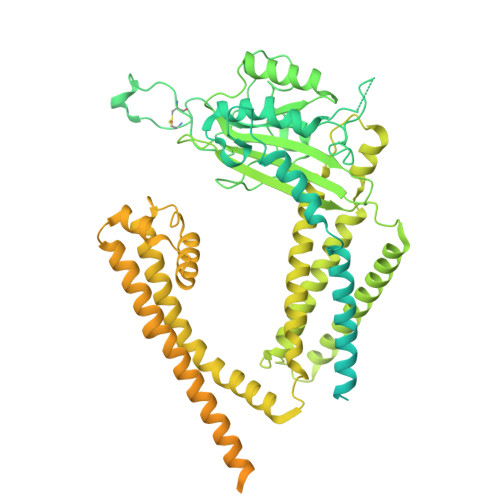 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13563 (Homo sapiens) Explore Q13563 Go to UniProtKB: Q13563 | |||||
PHAROS: Q13563 GTEx: ENSG00000118762 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13563 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1D75 (Subject of Investigation/LOI) Query on A1D75 | E [auth A] | [(2~{R})-2-hexadecanoyloxy-3-[oxidanyl-[(2~{R},3~{R},5~{S},6~{R})-2,3,5,6-tetrakis(oxidanyl)-4-phosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] hexadecanoate C41 H80 O16 P2 UJVUMTUBMCYKBK-SDTPXLKLSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 31930059 |
| National Natural Science Foundation of China (NSFC) | China | 81920108015 |