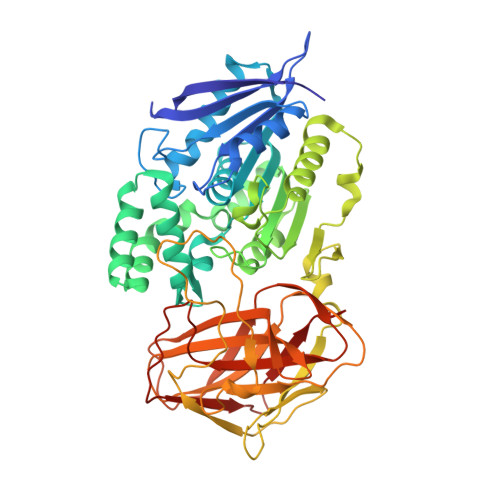
Crystal structure of lipase from Pseudomonas aeruginosa reveals an unusual catalytic triad conformation.
Xu, G., Guo, H., Yu, Z., Wang, S., Shen, D., Yang, L., Wu, J., Chen, B., Yu, H.(2024) Structure 32: 1454-1464.e3
- PubMed: 39025068
- DOI: https://doi.org/10.1016/j.str.2024.06.014
- Primary Citation of Related Structures:
8YZN, 8YZO - PubMed Abstract:
The Pseudomonas aeruginosa lipase PaL catalyzes the stereoselective hydrolysis of menthyl propionate to produce L-menthol. The lack of a three-dimensional structure of PaL has so far prevented a detailed understanding of its stereoselective reaction mechanism. Here, the crystal structure of PaL was determined at a resolution of 1.80 Å by single-wavelength anomalous diffraction. In the apo-PaL structure, the catalytic His302 is located in a long loop on the surface that is solvent exposed. His302 is distant from the other two catalytic residues, Asp274 and Ser164. This configuration of catalytic residues is unusual for lipases. Using metadynamics simulations, we observed that the enzyme undergoes a significant conformational change upon ligand binding. We also explored the catalytic and stereoselectivity mechanisms of PaL by all-atom molecular dynamics simulations. These findings could guide the engineering of PaL with an improved diastereoselectivity for L-menthol production.
- Institute of Bioengineering, College of Chemical and Biological Engineering, Zhejiang University, Hangzhou 310027, Zhejiang, China.
Organizational Affiliation: